 |
PDBsum entry 3n6r

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|


 591 a.a.
591 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 0 more)
506 a.a.
(+ 0 more)
506 a.a.
|
 |

|

|

|

|

|

|

|


 646 a.a.
646 a.a.
|
 |

|

|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
|
Structure:
|
 |
Propionyl-coa carboxylase, alpha subunit. Chain: a, c, e, g, i, k. Engineered: yes. Propionyl-coa carboxylase, beta subunit. Chain: b, d, f, h, j, l. Engineered: yes
|
|
Source:
|
 |
Ruegeria pomeroyi. Organism_taxid: 89184. Gene: pcca, spo1101. Expressed in: escherichia coli. Expression_system_taxid: 562. Roseobacter denitrificans. Organism_taxid: 375451. Strain: och 114. Gene: pccb, rd1_2028.
|
|
Resolution:
|
 |
|
3.20Å
|
R-factor:
|
0.212
|
R-free:
|
0.245
|
|
|
Authors:
|
 |
C.S.Huang,K.Sadre-Bazzaz,L.Tong
|
|
Key ref:
|
 |
C.S.Huang
et al.
(2010).
Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
Nature,
466,
1001-1005.
PubMed id:
|
 |
|
Date:
|
 |
|
26-May-10
|
Release date:
|
25-Aug-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5LUF3
(Q5LUF3_RUEPO) -
Propionyl-CoA carboxylase alpha chain from Ruegeria pomeroyi (strain ATCC 700808 / DSM 15171 / DSS-3)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
681 a.a.
591 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L:
E.C.6.4.1.3
- propionyl-CoA carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
propanoyl-CoA + hydrogencarbonate + ATP = (S)-methylmalonyl-CoA + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
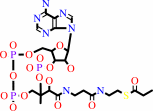 propanoyl-CoA
propanoyl-CoA
|
+
|
 hydrogencarbonate
hydrogencarbonate
|
+
|
 ATP
ATP
|
=
|
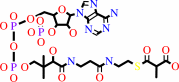 (S)-methylmalonyl-CoA
(S)-methylmalonyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin
|
 |
 |
 |
 |
 |
Biotin
Bound ligand (Het Group name =
BTI)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nature
466:1001-1005
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.
|
|
C.S.Huang,
K.Sadre-Bazzaz,
Y.Shen,
B.Deng,
Z.H.Zhou,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.S.Huang,
P.Ge,
Z.H.Zhou,
and
L.Tong
(2012).
An unanticipated architecture of the 750-kDa α6β6 holoenzyme of 3-methylcrotonyl-CoA carboxylase.
|
| |
Nature,
481,
219-223.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.A.Claridge,
J.J.Schwartz,
and
P.S.Weiss
(2011).
Electrons, photons, and force: quantitative single-molecule measurements from physics to biology.
|
| |
ACS Nano,
5,
693-729.
|
 |
|
|
|
|
 |
L.P.Yu,
Y.S.Kim,
and
L.Tong
(2010).
Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden.
|
| |
Proc Natl Acad Sci U S A,
107,
22072-22077.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
| |

