 |
PDBsum entry 3mp5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of human lyase r41m in complex with hmg-coa
|
|
Structure:
|
 |
Hydroxymethylglutaryl-coa lyase. Chain: a, b, c, d, e, f. Synonym: hmg-coa lyase, hl, 3-hydroxy-3-methylglutarate-coa lyase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: hmgcl. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.221
|
R-free:
|
0.271
|
|
|
Authors:
|
 |
Z.Fu,J.A.Runquist,C.Montgomery,H.M.Miziorko,J.-J.P.Kim
|
|
Key ref:
|
 |
Z.Fu
et al.
(2010).
Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J Biol Chem,
285,
26341-26349.
PubMed id:
|
 |
|
Date:
|
 |
|
24-Apr-10
|
Release date:
|
16-Jun-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P35914
(HMGCL_HUMAN) -
Hydroxymethylglutaryl-CoA lyase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
325 a.a.
296 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.3.4
- hydroxymethylglutaryl-CoA lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Mevalonate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(3S)-3-hydroxy-3-methylglutaryl-CoA = acetoacetate + acetyl-CoA
|
 |
 |
 |
 |
 |
(3S)-3-hydroxy-3-methylglutaryl-CoA
|
=
|
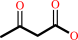 acetoacetate
acetoacetate
|
+
|
acetyl-CoA
Bound ligand (Het Group name = )
matches with 87.93% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Chem
285:26341-26349
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
|
|
Z.Fu,
J.A.Runquist,
C.Montgomery,
H.M.Miziorko,
J.J.Kim.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
HMG-CoA lyase (HMGCL) is crucial to ketogenesis and inherited human mutations
are potentially lethal. Detailed understanding of HMGCL reaction mechanism and
the molecular basis for correlating human mutations with enzyme deficiency have
been limited by the lack of structural information for enzyme liganded to an
acyl-CoA substrate or inhibitor. Crystal structures of ternary complexes of
wild-type HMGCL with the competitive inhibitor 3-hydroxyglutaryl-CoA (HG-CoA)
and of the catalytically deficient HMGCL R41M mutant with substrate HMG-CoA have
been determined to 2.4 A and 2.2 A,respectively. Comparison of these beta/alpha
barrel structures with those of unliganded HMGCL and R41M reveals substantial
differences for Mg++ coordination and positioning of the flexible loop
containing the conserved HMGCL signature sequence. In the ternary
substrate-Mg++-R41M complex, loop residue C266 (implicated in active site
function by mechanistic and mutagenesis observations) is more closely juxtaposed
to the catalytic site than in the case of unliganded enzyme or the wild-type
enzyme-Mg++-HG-CoA inhibitor complex. In both ternary complexes, the
S-stereoisomer of substrate or inhibitor is specifically bound, in accord with
the observed Mg++ liganding of both C3 hydroxyl and C5 carboxyl oxygens. In
addition to H233 and H235 imidazoles, other Mg++ ligands are D42 carboxyl oxygen
and an ordered water molecule. This water, positioned between D42 and the C3
hydroxyl of bound substrate/inhibitor, may function as a proton shuttle. The
observed interaction of R41 with acyl-CoA C1 carbonyl oxygen explains the
effects of R41 mutation on reaction product enolization and accounts for why
human R41 mutations cause drastic enzyme deficiency.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

