 |
PDBsum entry 3lp7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.3.1
- arginase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arginine + H2O = urea + L-ornithine
|
 |
 |
 |
 |
 |
 L-arginine
L-arginine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 92.31% similarity
|
=
|
 urea
urea
|
+
|
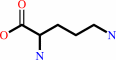 L-ornithine
L-ornithine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Arch Biochem Biophys
496:101-108
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibition of human arginase I by substrate and product analogues.
|
|
L.Di Costanzo,
M.Ilies,
K.J.Thorn,
D.W.Christianson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human arginase I is a binuclear manganese metalloenzyme that catalyzes the
hydrolysis of L-arginine to generate L-ornithine and urea. We demonstrate that
N-hydroxy-L-arginine (NOHA) binds to this enzyme with K(d)=3.6 microM, and
nor-N-hydroxy-L-arginine (nor-NOHA) binds with K(d)=517 nM (surface plasmon
resonance) or K(d) approximately 50 nM (isothermal titration calorimetry).
Crystals of human arginase I complexed with NOHA and nor-NOHA afford 2.04 and
1.55 A resolution structures, respectively, which are significantly improved in
comparison with previously-determined structures of the corresponding complexes
with rat arginase I. Higher resolution structures clarify the binding
interactions of the inhibitors. Finally, the crystal structure of the complex
with L-lysine (K(d)=13 microM) is reported at 1.90 A resolution. This structure
confirms the importance of hydrogen bond interactions with inhibitor
alpha-carboxylate and alpha-amino groups as key specificity determinants of
amino acid recognition in the arginase active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Riley,
S.C.Roberts,
and
B.Ullman
(2011).
Inhibition profile of Leishmania mexicana arginase reveals differences with human arginase I.
|
| |
Int J Parasitol,
41,
545-552.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

