 |
PDBsum entry 3ln6
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of a bifunctional glutathione synthetase from streptococcus agalactiae
|
|
Structure:
|
 |
Glutathione biosynthesis bifunctional protein gshab. Chain: a. Synonym: gamma-gcs-gs, gcs-gs, glutamate-cysteine ligase, gamma- glutamylcysteine synthetase, gamma-ecs, gcs, glutathione synthetase, glutathione synthase, gsh synthetase, gsh-s, gshase, gs. Engineered: yes
|
|
Source:
|
 |
Streptococcus agalactiae serogroup v. Organism_taxid: 216466. Strain: baa-811. Gene: gshab, gshf, sag1821. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.95Å
|
R-factor:
|
0.253
|
R-free:
|
0.286
|
|
|
Authors:
|
 |
J.Stout,B.Vergauwen,S.N.Savvides
|
|
Key ref:
|
 |
J.Stout
et al.
Structures of two bifunctional gamma-Glutamate-Cystei ligase/glutathione synthetases (gshf) reveal a novel ATP-Grasp fold.
To be published,
.
|
 |
|
Date:
|
 |
|
02-Feb-10
|
Release date:
|
13-Apr-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8DXM9
(GSHAB_STRA5) -
Glutathione biosynthesis bifunctional protein GshAB from Streptococcus agalactiae serotype V (strain ATCC BAA-611 / 2603 V/R)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
750 a.a.
743 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.6.3.2.2
- glutamate--cysteine ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cysteine + L-glutamate + ATP = gamma-L-glutamyl-L-cysteine + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 L-cysteine
L-cysteine
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ATP
ATP
|
=
|
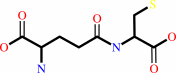 gamma-L-glutamyl-L-cysteine
gamma-L-glutamyl-L-cysteine
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.3.2.3
- glutathione synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
gamma-L-glutamyl-L-cysteine + glycine + ATP = glutathione + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 gamma-L-glutamyl-L-cysteine
gamma-L-glutamyl-L-cysteine
|
+
|
 glycine
glycine
|
+
|
 ATP
ATP
|
=
|
 glutathione
glutathione
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

