 |
PDBsum entry 3ipi

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.81
- geranylfarnesyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + (2E,6E,10E)-geranylgeranyl diphosphate = (2E,6E,10E,14E)-geranylfarnesyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
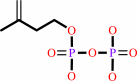 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
(2E,6E,10E)-geranylgeranyl diphosphate
|
=
|
(2E,6E,10E,14E)-geranylfarnesyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

