 |
PDBsum entry 3gos

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate n- succinyltransferase from yersinia pestis co92
|
|
Structure:
|
 |
2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n- succinyltransferase. Chain: a, b, c. Synonym: tetrahydrodipicolinate n-succinyltransferase, thp succinyltransferase, tetrahydropicolinate succinylase. Engineered: yes
|
|
Source:
|
 |
Yersinia pestis. Organism_taxid: 632. Strain: co92. Gene: dapd, gi: 16121341, y3140, ypo1041, yp_2810. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.230
|
R-free:
|
0.272
|
|
|
Authors:
|
 |
R.Zhang,N.Maltseva,K.Kwon,W.Anderson,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
R.Zhang
et al.
The crystal structure of 2,3,4,5-Tetrahydropyridine-2-Carboxylate n-Succinyltransferase from yersinia pestis co92.
To be published,
.
|
 |
|
Date:
|
 |
|
19-Mar-09
|
Release date:
|
12-May-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8ZH69
(DAPD_YERPE) -
2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Yersinia pestis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
274 a.a.
270 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.117
- 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Lysine biosynthesis (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-2,3,4,5-tetrahydrodipicolinate + succinyl-CoA + H2O = (S)-2- succinylamino-6-oxoheptanedioate + CoA
|
 |
 |
 |
 |
 |
(S)-2,3,4,5-tetrahydrodipicolinate
|
+
|
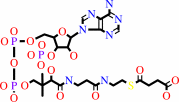 succinyl-CoA
succinyl-CoA
|
+
|
H2O
|
=
|
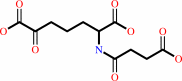 (S)-2- succinylamino-6-oxoheptanedioate
(S)-2- succinylamino-6-oxoheptanedioate
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

