 |
PDBsum entry 3elp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.4.1.1
- cystathionine gamma-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L,L-cystathionine + H2O = 2-oxobutanoate + L-cysteine + NH4+
|
 |
 |
 |
 |
 |
L,L-cystathionine
|
+
|
H2O
|
=
|
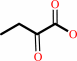 2-oxobutanoate
2-oxobutanoate
|
+
|
 L-cysteine
L-cysteine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.2
- homocysteine desulfhydrase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homocysteine + H2O = 2-oxobutanoate + hydrogen sulfide + NH4+ + H+
|
 |
 |
 |
 |
 |
 L-homocysteine
L-homocysteine
|
+
|
H2O
|
=
|
 2-oxobutanoate
2-oxobutanoate
|
+
|
hydrogen sulfide
|
+
|
NH4(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
284:3076-3085
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S.
|
|
Q.Sun,
R.Collins,
S.Huang,
L.Holmberg-Schiavone,
G.S.Anand,
C.H.Tan,
S.van-den-Berg,
L.W.Deng,
P.K.Moore,
T.Karlberg,
J.Sivaraman.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Impairment of the formation or action of hydrogen sulfide (H(2)S), an endogenous
gasotransmitter, is associated with various diseases, such as hypertension,
diabetes mellitus, septic and hemorrhagic shock, and pancreatitis. Cystathionine
beta-synthase and cystathionine gamma-lyase (CSE) are two pyridoxal-5'-phosphate
(PLP)-dependent enzymes largely responsible for the production of H(2)S in
mammals. Inhibition of CSE by DL-propargylglycine (PAG) has been shown to
alleviate disease symptoms. Here we report crystal structures of human CSE
(hCSE), in apo form, and in complex with PLP and PLP.PAG. Structural
characterization, combined with biophysical and biochemical studies, provides
new insights into the inhibition mechanism of hCSE-mediated production of H(2)S.
Transition from the open form of apo-hCSE to the closed PLP-bound form reveals
large conformational changes hitherto not reported. In addition, PAG binds hCSE
via a unique binding mode, not observed in PAG-enzyme complexes previously. The
interaction of PAG-hCSE was not predicted based on existing information from
known PAG complexes. The structure of hCSE.PLP.PAG complex highlights the
particular importance of Tyr(114) in hCSE and the mechanism of PAG-dependent
inhibition of hCSE. These results provide significant insights, which will
facilitate the structure-based design of novel inhibitors of hCSE to aid in the
development of therapies for diseases involving disorders of sulfur metabolism.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Crystal structure of hCSE. a, ribbon diagram of the hCSE
monomer. N and C termini are labeled. The large PLP binding
domain is shown in red, the small domain in green, and PLP in
yellow. Panels b and c are the superimposed apo-hCSE (blue) and
PLP complex (red) active site regions. The Met^110–Asn^118
region (b) and loop Thr^211–Met^214 (c) are shown. The root
mean square deviation between two superimposed models is 1.5
Å for 336 Cα atoms. PLP, Tyr^114, and Lys^212 are shown
in stick representations. d, stereoview of the 2F[o] - F[c]
simulated annealing omit map of PLP from hCSE·PLP. All
atoms within 3.5 Å of PLP were omitted prior to
refinement. The map was contoured at a level of 1.0σ. These
figures were prepared using the program PyMol (32). Shown is the
electrostatic surface potential at the active site region of
apo-hCSE (e) and hCSE·PLP (f). Apo-hCSE shows significant
enlarged (open) surface. These figures were prepared using the
program GRASP (33).
|
 |
Figure 5.
Mechanism of PAG inhibition on hCSE. Upon the addition of the
inhibitor, the α-amino group of PAG is first deprotonated by
Arg^62 of the adjacent monomer (B:) (Step 1) for
transaldimination to occur (Step 2). Lys^212 then abstracts a
proton from the β-position of the bound alkyne to generate an
activated allene (Step 3), which is then attacked by the
hydroxyl group of Tyr^114 (Step 4) to produce a vinyl ether.
Subsequent transaldimination with Lys^212 (Step 5) regenerates
the internal aldimine.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2009,
284,
3076-3085)
copyright 2009.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.R.Linden,
M.D.Levitt,
G.Farrugia,
and
J.H.Szurszewski
(2010).
Endogenous production of H2S in the gastrointestinal tract: still in search of a physiologic function.
|
| |
Antioxid Redox Signal,
12,
1135-1146.
|
 |
|
|
|
|
 |
A.F.Perna,
M.G.Luciano,
D.Ingrosso,
P.Pulzella,
I.Sepe,
D.Lanza,
E.Violetti,
R.Capasso,
C.Lombardi,
and
N.G.De Santo
(2009).
Hydrogen sulphide-generating pathways in haemodialysis patients: a study on relevant metabolites and transcriptional regulation of genes encoding for key enzymes.
|
| |
Nephrol Dial Transplant,
24,
3756-3763.
|
 |
|
|
|
|
 |
L.Li,
A.Hsu,
and
P.K.Moore
(2009).
Actions and interactions of nitric oxide, carbon monoxide and hydrogen sulphide in the cardiovascular system and in inflammation--a tale of three gases!
|
| |
Pharmacol Ther,
123,
386-400.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

