 |
PDBsum entry 3e2t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3e2t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.16.4
- tryptophan 5-monooxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biopterin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin + L-tryptophan + O2 = 5-hydroxy-L-tryptophan + (4aS,6R)-4a-hydroxy-L-erythro-5,6,7,8- tetrahydrobiopterin
|
 |
 |
 |
 |
 |
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin
|
+
|
L-tryptophan
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O2
O2
|
=
|
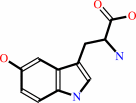 5-hydroxy-L-tryptophan
5-hydroxy-L-tryptophan
|
+
|
(4aS,6R)-4a-hydroxy-L-erythro-5,6,7,8- tetrahydrobiopterin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:12087-12094
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of tryptophan hydroxylase with bound amino acid substrate.
|
|
M.S.Windahl,
C.R.Petersen,
H.E.Christensen,
P.Harris.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tryptophan hydroxylase (TPH) is a mononuclear non-heme iron enzyme, which
catalyzes the reaction between tryptophan, O 2, and tetrahydrobiopterin (BH 4)
to produce 5-hydroxytryptophan and 4a-hydroxytetrahydrobiopterin. This is the
first and rate-limiting step in the biosynthesis of the neurotransmitter and
hormone serotonin (5-hydroxytryptamine). We have determined the 1.9 A resolution
crystal structure of the catalytic domain (Delta1-100/Delta415-445) of chicken
TPH isoform 1 (TPH1) in complex with the tryptophan substrate and an iron-bound
imidazole. This is the first structure of any aromatic amino acid hydroxylase
with bound natural amino acid substrate. The iron coordination can be described
as distorted trigonal bipyramidal coordination with His273, His278, and Glu318
(partially bidentate) and one imidazole as ligands. The tryptophan stacks
against Pro269 with a distance of 3.9 A between the iron and the tryptophan
Czeta3 atom that is hydroxylated. The binding of tryptophan and maybe the
imidazole has caused the structural changes in the catalytic domain compared to
the structure of the human TPH1 without tryptophan. The structure of chicken
TPH1 is more compact, and the loops of residues Leu124-Asp139 and Ile367-Thr369
close around the active site. Similar structural changes are seen in the
catalytic domain of phenylalanine hydroxylase (PAH) upon binding of substrate
analogues norleucine and thienylalanine to the PAH.BH 4 complex. In fact, the
chicken TPH1.Trp.imidazole structure resembles the PAH.BH 4.thienylalanine
structure more (root-mean-square deviation for Calpha atoms of 0.90 A) than the
human TPH1 structure (root-mean-square deviation of 1.47 A).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Olsson,
A.Martinez,
K.Teigen,
and
V.R.Jensen
(2011).
Formation of the iron-oxo hydroxylating species in the catalytic cycle of aromatic amino acid hydroxylases.
|
| |
Chemistry,
17,
3746-3758.
|
 |
|
|
|
|
 |
V.Helmetag,
S.A.Samel,
M.G.Thomas,
M.A.Marahiel,
and
L.O.Essen
(2009).
Structural basis for the erythro-stereospecificity of the L-arginine oxygenase VioC in viomycin biosynthesis.
|
| |
FEBS J,
276,
3669-3682.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

