 |
PDBsum entry 3dp9

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.2.9
- adenosylhomocysteine nucleosidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Autoinducer AI-2 Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
S-adenosyl-L-homocysteine + H2O = S-(5-deoxy-D-ribos-5-yl)-L- homocysteine + adenine
|
|
2.
|
5'-deoxyadenosine + H2O = 5-deoxy-D-ribose + adenine
|
|
3.
|
S-methyl-5'-thioadenosine + H2O = 5-(methylsulfanyl)-D-ribose + adenine
|
|
 |
 |
 |
 |
 |
 S-adenosyl-L-homocysteine
S-adenosyl-L-homocysteine
|
+
|
H2O
|
=
|
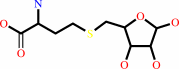 S-(5-deoxy-D-ribos-5-yl)-L- homocysteine
S-(5-deoxy-D-ribos-5-yl)-L- homocysteine
|
+
|
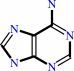 adenine
adenine
|
|
 |
 |
 |
 |
 |
5'-deoxyadenosine
Bound ligand (Het Group name = )
matches with 41.38% similarity
|
+
|
H2O
|
=
|
5-deoxy-D-ribose
|
+
|
 adenine
adenine
|
|
 |
 |
 |
 |
 |
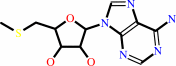 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
H2O
|
=
|
5-(methylsulfanyl)-D-ribose
|
+
|
 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
5:251-257
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Transition state analogs of 5'-methylthioadenosine nucleosidase disrupt quorum sensing.
|
|
J.A.Gutierrez,
T.Crowder,
A.Rinaldo-Matthis,
M.C.Ho,
S.C.Almo,
V.L.Schramm.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
5'-Methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN) is a bacterial
enzyme involved in S-adenosylmethionine-related quorum sensing pathways that
induce bacterial pathogenesis factors. Transition state analogs
MT-DADMe-Immucillin-A, EtT-DADMe-Immucillin-A and BuT-DADMe-Immucillin-A are
slow-onset, tight-binding inhibitors of Vibrio cholerae MTAN (VcMTAN), with
equilibrium dissociation constants of 73, 70 and 208 pM, respectively.
Structural analysis of VcMTAN with BuT-DADMe-Immucillin-A revealed interactions
contributing to the high affinity. We found that in V. cholerae cells, these
compounds are potent MTAN inhibitors with IC(50) values of 27, 31 and 6 nM for
MT-, EtT- and BuT-DADMe-Immucillin-A, respectively; the compounds disrupt
autoinducer production in a dose-dependent manner without affecting growth. MT-
and BuT-DADMe-Immucillin-A also inhibited autoinducer-2 production in
enterohemorrhagic Escherichia coli O157:H7 with IC(50) values of 600 and 125 nM,
respectively. BuT-DADMe-Immucillin-A inhibition of autoinducer-2 production in
both strains persisted for several generations and caused reduction in biofilm
formation. These results support MTAN's role in quorum sensing and its potential
as a target for bacterial anti-infective drug design.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
(a) Top, a dissociative transition state for E. coli with
ribooxacarbenium ion character^10. Bottom, structures of stable
analogs for an early dissociative transition state (ImmA) and a
late dissociative transition state (DADMe-ImmA) depict
differences in bond distances between the adenine leaving group
and the ribosyl group, as well as charge localization. TS,
transition state. (b) The structure of S-substituted DADMe-ImmA,
along with MT, EtT and BuT substituents.
|
 |
Figure 3.
(a) Overall structure of VcMTAN showing the asymmetric unit
content with the inhibitor BuT-DADMe-ImmA bound in the active
sites. (b) The active site of VcMTAN with a 2F[o] - F[c] map
contoured at 1.2  surrounding
the BuT-DADMe-ImmA inhibitor and the proposed nucleophilic water
molecule. (c) Space-filling picture of the active site of VcMTAN
with BuT-DADMe-ImmA in the active site. Grey represents
hydrophobic regions of the protein, which interact with
hydrophobic parts of the inhibitor. The red color shows parts of
the protein that contain charged residues interacting with polar
groups of the inhibitor, and green represents loop regions. (d)
Schematic drawing of the BuT-DADMe-ImmA inhibitor bound in the
active site of VcMTAN showing catalytic contacts. surrounding
the BuT-DADMe-ImmA inhibitor and the proposed nucleophilic water
molecule. (c) Space-filling picture of the active site of VcMTAN
with BuT-DADMe-ImmA in the active site. Grey represents
hydrophobic regions of the protein, which interact with
hydrophobic parts of the inhibitor. The red color shows parts of
the protein that contain charged residues interacting with polar
groups of the inhibitor, and green represents loop regions. (d)
Schematic drawing of the BuT-DADMe-ImmA inhibitor bound in the
active site of VcMTAN showing catalytic contacts.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
Nat Chem Biol
(2009,
5,
251-257)
copyright 2009.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Fernebro
(2011).
Fighting bacterial infections-Future treatment options.
|
| |
Drug Resist Updat,
14,
125-139.
|
 |
|
|
|
|
 |
N.Parveen,
and
K.A.Cornell
(2011).
Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism.
|
| |
Mol Microbiol,
79,
7.
|
 |
|
|
|
|
 |
D.R.Ronning,
N.M.Iacopelli,
and
V.Mishra
(2010).
Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
|
| |
Protein Sci,
19,
2498-2510.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Augustine,
P.Kumar,
and
S.Thomas
(2010).
Inhibition of Vibrio cholerae biofilm by AiiA enzyme produced from Bacillus spp.
|
| |
Arch Microbiol,
192,
1019-1022.
|
 |
|
|
|
|
 |
S.D.Schwartz,
and
V.L.Schramm
(2009).
Enzymatic transition states and dynamic motion in barrier crossing.
|
| |
Nat Chem Biol,
5,
551-558.
|
 |
|
|
|
|
 |
V.L.Schramm
(2009).
Transition States.
|
| |
J Biol Chem,
284,
32201-32208.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

