 |
PDBsum entry 3acl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Metal binding protein
|
PDB id
|
|

|

|
3acl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.24
- quercetin 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Quercetin 2,3-Dioxygenase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
quercetin + O2 = 2-(3,4-dihydroxybenzoyloxy)-4,6-dihydroxybenzoate + CO
|
 |
 |
 |
 |
 |
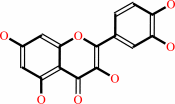 quercetin
quercetin
|
+
|
O2
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
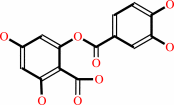 2-(3,4-dihydroxybenzoyloxy)-4,6-dihydroxybenzoate
2-(3,4-dihydroxybenzoyloxy)-4,6-dihydroxybenzoate
|
+
|
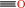 CO
CO
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation or Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Chem Biol
6:667-673
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
A small-molecule inhibitor shows that pirin regulates migration of melanoma cells.
|
|
I.Miyazaki,
S.Simizu,
H.Okumura,
S.Takagi,
H.Osada.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

