 |
PDBsum entry 3a2n

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.8.1.5
- haloalkane dehalogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-haloalkane + H2O = a halide anion + a primary alcohol + H+
|
 |
 |
 |
 |
 |
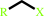 1-haloalkane
1-haloalkane
|
+
|
H2O
|
=
|
halide anion
|
+
|
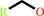 primary alcohol
primary alcohol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Angew Chem Int Ed Engl
49:6111-6115
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering.
|
|
Z.Prokop,
Y.Sato,
J.Brezovsky,
T.Mozga,
R.Chaloupkova,
T.Koudelakova,
P.Jerabek,
V.Stepankova,
R.Natsume,
J.G.van Leeuwen,
D.B.Janssen,
J.Florian,
Y.Nagata,
T.Senda,
J.Damborsky.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

