 |
PDBsum entry 2zz1

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
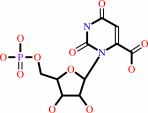 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 88.00% similarity
|
=
|
 UMP
UMP
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 95.45% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
387:1199-1210
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural characterization of the molecular events during a slow substrate-product transition in orotidine 5'-monophosphate decarboxylase.
|
|
M.Fujihashi,
L.Wei,
L.P.Kotra,
E.F.Pai.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystal structures of substrate-product complexes of Methanobacterium
thermoautotrophicum orotidine 5'-monophosphate decarboxylase, obtained at
various steps in its catalysis of the unusual transformation of 6-cyano-uridine
5'-monophosphate (UMP) into barbituric acid ribosyl monophosphate, show that the
cyano substituent of the substrate, when bound to the active site, is first bent
significantly from the plane of the pyrimidine ring and then replaced by an
oxygen atom. Although the K72A and D70A/K72A mutants are either catalytically
impaired or even completely inactive, they still display bending of the C6
substituent. Interestingly, high-resolution structures of the D70A and D75N
mutants revealed a covalent bond between C6 of UMP and the Lys72 side chain
after the -CN moiety's release. The same covalent bond was observed when the
native enzyme was incubated with 6-azido-UMP and 6-iodo-UMP; in contrast, the
K72A mutant transformed 6-iodo-UMP to barbituric acid ribosyl 5'-monophosphate.
These results demonstrate that, given a suitable environment, native orotidine
5'-monophosphate decarboxylase and several of its mutants are not restricted to
the physiologically relevant decarboxylation; they are able to catalyze even
nucleophilic substitution reactions but consistently maintain distortion on the
C6 substituent as an important feature of catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Close-ups of the active sites of native and mutant
MtODCase complexes with 6-CN-UMP. (a–d) F[o] − F[c] omit
electron density maps superimposed on the atomic model. The
pyrimidine ring and residues Lys72, Asp70, Lys42, and Asp75^B
(or the corresponding mutant residues) are omitted for the map
calculation. (a) K72A mutant. Map contoured at 2.5σ. (b) Native
MtODCase incubated with 6-CN-UMP for 1 day at RT and 1 day at 4
°C. (c) Native MtODCase incubated with 6-CN-UMP for 2 months
at RT. (d) D70A/K72A double mutant. Maps (b)–(d) are contoured
at 3.0σ. (e) View approximately perpendicular to that in (b).
The F[o] − F[c] omit electron density map was calculated with
the pyrimidine ring and the surrounding water molecules excluded
from the phasing model. (f) F[o](2 days) − F[o](2 months)
difference electron density map superimposed on the model of the
native MtODCase–6-CN-UMP complex. The ligand molecule was
excluded from the model used in calculating the phases for this
map. Green (positive difference density) and red (negative
difference density) meshes are contoured at + 6.5σ and −
6.5σ, respectively. (g–j) Stereo views of the side-chain
arrangements in the reaction center of MtODCase. Numbers colored
in both blue and red indicate the distances between atoms in
Ångstrom. (g) The K72A–6-CN-UMP complex. Lys72 of native
MtODCase is superimposed for comparison and shown in transparent
gray. (h) Native MtODCase–6-CN-UMP incubated for 1 day at RT
and 1 day at 4 °C. The hydroxyl group of the product BMP is
shown in transparent gray and red. (i) D70A/K72A–6-CN-UMP.
Asp70 and Lys72 of native ODCase are superimposed for comparison
and shown in transparent gray. (j) D70A/K72A–OMP (PDB ID
1KM6). The D70A/K72A–6-CN-UMP complex was superimposed, and
6-CN-UMP is shown in transparent gray for comparison. Water
molecules bound in the D70A/K72A–6-CN-UMP complex are shown in
green. These panels were prepared with PyMOL
(http://pymol.sourceforge.net).
|
 |
Figure 6.
Fig. 6. Cartoon of the proposed reaction schemes. Distortions
of the carboxyl, cyano, iodo, and azido groups play an important
role in the reaction catalyzed by ODCase. “P-Rib” indicates
ribose 5′-monophosphate. (a) The reaction from OMP to UMP
catalyzed by native ODCase. (b) Covalent-bond formation between
the mutant ODCase and 6-CN-UMP. (c) The conversion of 6-CN-UMP
to BMP catalyzed by native ODCase. (d) Covalent-bond formation
between the native ODCase and 6-I-UMP or 6-N[3]-UMP. (e) The
conversion of 6-I-UMP to BMP by K72A MtODCase. Distortion of the
bond linking the pyrimidine ring to the C6 substituent is
conserved in all these reactions, the physiologically relevant
decarboxylation and the nucleophilic mechanisms.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2009,
387,
1199-1210)
copyright 2009.
|
|
| |
Figures were
selected
by the author.
|
|

|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
The results of our "slow motion" time-resolved crystallography (enabled by the very slow reaction rates of the 6-CN-UMP substrate analogue) on native orotidine 5'-monophosphate decarboxylase (ODCase) show that the enzyme employs distortion on the C6 substituent as an important feature in its catalysis. Additional structures of complexes of ODCase mutants with a variety of C6-substituted substrate analogues demonstrate that, given a suitable environment, the enzyme is not restricted to the physiologically relevant decarboxylation but is able to catalyze even nucleophilic substitutions.
Masahiro Fujihashi, Emil F. Pai
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.J.Wu,
C.C.Liao,
C.H.Jen,
Y.C.Shih,
and
T.C.Chien
(2010).
Chemical models and their mechanistic implications for the transformation of 6-cyanouridine 5'-monophosphate catalyzed by orotidine 5'-monophosphate decarboxylase.
|
| |
Chem Commun (Camb),
46,
4821-4823.
|
 |
|
|
|
|
 |
D.Heinrich,
U.Diederichsen,
and
M.G.Rudolph
(2009).
Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase.
|
| |
Chemistry,
15,
6619-6625.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

