 |
PDBsum entry 2yzd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2yzd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of uricase from arthrobacter globiformis in complex with 8-azaxanthin (inhibitor)
|
|
Structure:
|
 |
Uricase. Chain: a, b, c, d, e, f, g, h. Ec: 1.7.3.3
|
|
Source:
|
 |
Arthrobacter globiformis
|
|
Resolution:
|
 |
|
2.24Å
|
R-factor:
|
0.189
|
R-free:
|
0.239
|
|
|
Authors:
|
 |
E.C.M.Juan,M.T.Hossain,M.M.Hoque,K.Suzuki,T.Sekiguchi,A.Takenaka
|
|
Key ref:
|
 |
E.C.M.Juan
et al.
Trapping of the uric acid substrate in the crystal structure of urate oxidase from arthrobacter globiformis.
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
05-May-07
|
Release date:
|
06-May-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
D0VWQ1
(URIC_ARTGO) -
Uricase from Arthrobacter globiformis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
302 a.a.
287 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.3.3
- factor independent urate hydroxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
urate + O2 + H2O = 5-hydroxyisourate + H2O2
|
 |
 |
 |
 |
 |
urate
Bound ligand (Het Group name = )
matches with 76.92% similarity
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
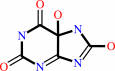 5-hydroxyisourate
5-hydroxyisourate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Copper
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

