 |
PDBsum entry 2xo4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2xo4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Ribonucleotide reductase y730nh2y modified r1 subunit of e. Coli
|
|
Structure:
|
 |
Ribonucleoside-diphosphate reductase 1 subunit alpha. Chain: a, b, c. Fragment: residues 1-761. Synonym: ribonucleoside-diphosphate reductase 1 r1 subunit, ribonucleotide reductase 1, protein b1, ribonucleotide reductase r1 subunit. Engineered: yes. Other_details: site specific incorporation of 3-aminotyrosine at position 730.
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 83333. Strain: k-12. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Organism_taxid: 562
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.191
|
R-free:
|
0.233
|
|
|
Authors:
|
 |
E.C.Minnihan,M.R.Seyedsayamdost,U.Uhlin,J.Stubbe
|
|
Key ref:
|
 |
E.C.Minnihan
et al.
(2011).
Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases.
J Am Chem Soc,
133,
9430-9440.
PubMed id:
|
 |
|
Date:
|
 |
|
09-Aug-10
|
Release date:
|
18-Aug-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
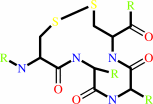 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
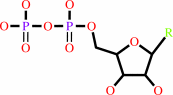 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
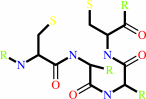 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
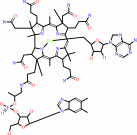 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Am Chem Soc
133:9430-9440
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetics of radical intermediate formation and deoxynucleotide production in 3-aminotyrosine-substituted Escherichia coli ribonucleotide reductases.
|
|
E.C.Minnihan,
M.R.Seyedsayamdost,
U.Uhlin,
J.Stubbe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

