 |
PDBsum entry 2wu8

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.9
- glucose-6-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucose 6-phosphate = beta-D-fructose 6-phosphate
|
 |
 |
 |
 |
 |
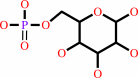 alpha-D-glucose 6-phosphate
alpha-D-glucose 6-phosphate
|
=
|
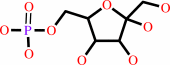 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
66:490-497
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies of phosphoglucose isomerase from Mycobacterium tuberculosis H37Rv.
|
|
K.Anand,
D.Mathur,
A.Anant,
L.C.Garg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphoglucose isomerase (PGI) plays a key role in both glycolysis and
gluconeogenesis inside the cell, whereas outside the cell it exhibits cytokine
properties. PGI is also known to act as an autocrine motility factor, a
neuroleukin agent and a differentiation and maturation mediator. Here, the first
crystal structure of PGI from Mycobacterium tuberculosis H37Rv (Mtb) is
reported. The structure was refined at 2.25 A resolution and revealed the
presence of one molecule in the asymmetric unit with two globular domains. As
known previously, the active site of Mtb PGI contains conserved residues
including Glu356, Glu216 and His387 (where His387 is from the neighbouring
molecule). The crystal structure of Mtb PGI was observed to be rather more
similar to human PGI than other nonbacterial PGIs, with only a few differences
being detected in the loops, arm and hook regions of the human and Mtb PGIs,
suggesting that the M. tuberculosis enzyme uses the same enzyme mechanism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

