 |
PDBsum entry 2wpe

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2wpe
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (ddd00073359)
|
|
Structure:
|
 |
Trypanothione reductase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Trypanosoma brucei. Organism_taxid: 185431. Strain: treu927. Expressed in: escherichia coli. Expression_system_taxid: 469008. Expression_system_variant: codonplus ril.
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.171
|
R-free:
|
0.230
|
|
|
Authors:
|
 |
M.S.Alphey,S.Patterson,A.H.Fairlamb
|
|
Key ref:
|
 |
S.Patterson
et al.
(2011).
Dihydroquinazolines as a novel class of Trypanosoma brucei trypanothione reductase inhibitors: discovery, synthesis, and characterization of their binding mode by protein crystallography.
J Med Chem,
54,
6514-6530.
PubMed id:
|
 |
|
Date:
|
 |
|
06-Aug-09
|
Release date:
|
13-Oct-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q389T8
(Q389T8_TRYB2) -
Trypanothione reductase from Trypanosoma brucei brucei (strain 927/4 GUTat10.1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
492 a.a.
490 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.12
- trypanothione-disulfide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
trypanothione + NADP+ = trypanothione disulfide + NADPH + H+
|
 |
 |
 |
 |
 |
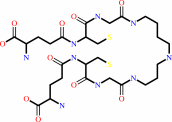 trypanothione
trypanothione
|
+
|
 NADP(+)
NADP(+)
|
=
|
 trypanothione disulfide
trypanothione disulfide
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Med Chem
54:6514-6530
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Dihydroquinazolines as a novel class of Trypanosoma brucei trypanothione reductase inhibitors: discovery, synthesis, and characterization of their binding mode by protein crystallography.
|
|
S.Patterson,
M.S.Alphey,
D.C.Jones,
E.J.Shanks,
I.P.Street,
J.A.Frearson,
P.G.Wyatt,
I.H.Gilbert,
A.H.Fairlamb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

