 |
PDBsum entry 2vlh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.99.2
- tyrosine phenol-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tyrosine + H2O = phenol + pyruvate + NH4+
|
 |
 |
 |
 |
 |
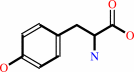 L-tyrosine
L-tyrosine
|
+
|
H2O
|
=
|
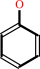 phenol
phenol
|
+
|
pyruvate
Bound ligand (Het Group name = )
matches with 46.67% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PM9)
matches with 60.00% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:29206-29214
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Insights into the catalytic mechanism of tyrosine phenol-lyase from X-ray structures of quinonoid intermediates.
|
|
D.Milić,
T.V.Demidkina,
N.G.Faleev,
D.Matković-Calogović,
A.A.Antson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Amino acid transformations catalyzed by a number of pyridoxal 5'-phosphate
(PLP)-dependent enzymes involve abstraction of the Calpha proton from an
external aldimine formed between a substrate and the cofactor leading to the
formation of a quinonoid intermediate. Despite the key role played by the
quinonoid intermediates in the catalysis by PLP-dependent enzymes, limited
accurate information is available about their structures. We trapped the
quinonoid intermediates of Citrobacter freundii tyrosine phenol-lyase with
L-alanine and L-methionine in the crystalline state and determined their
structures at 1.9- and 1.95-A resolution, respectively, by cryo-crystallography.
The data reveal a network of protein-PLP-substrate interactions that stabilize
the planar geometry of the quinonoid intermediate. In both structures the
protein subunits are found in two conformations, open and closed, uncovering the
mechanism by which binding of the substrate and restructuring of the active site
during its closure protect the quinonoid intermediate from the solvent and bring
catalytically important residues into positions suitable for the abstraction of
phenol during the beta-elimination of L-tyrosine. In addition, the structural
data indicate a mechanism for alanine racemization involving two bases, Lys-257
and a water molecule. These two bases are connected by a hydrogen bonding system
allowing internal transfer of the Calpha proton.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
TPL tetramer. Ribbon diagram with different subunits shown in
different colors. One catalytic dimer consists of the subunits
shown in cyan and blue, while the other is formed from the
subunits shown in orange and yellow. PLP and the side chains of
the PLP-binding Lys-257 residues are shown as spheres. The view
is along the crystallographic 2-fold axis.
|
 |
Figure 3.
Enzyme interactions with the Ala quinonoid intermediate in
the closed (A) and open (B) active sites. Stereo views with
structures of quinonoid and two water molecules superposed with
the corresponding weighted |F[o]| - |F[c]| electron density omit
maps (green) are contoured at the 3.0σ level. Hydrogen bonds
are denoted by dashed lines. Carbon atoms of residues belonging
to the large rigid region are shown in orange, those from the
small rigid region are shown in pink, and the residues from the
neighboring subunit are shown in blue and labeled with a star.
The alternate conformations of Thr-124, Thr-216, Met-288, and
Phe-449 in the open conformation are shown in corresponding pale
tones.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the ASBMB:
J Biol Chem
(2008,
283,
29206-29214)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Koutmos,
O.Kabil,
J.L.Smith,
and
R.Banerjee
(2010).
Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine {beta}-synthase.
|
| |
Proc Natl Acad Sci U S A,
107,
20958-20963.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Rha,
S.Kim,
S.L.Choi,
S.P.Hong,
M.H.Sung,
J.J.Song,
and
S.G.Lee
(2009).
Simultaneous improvement of catalytic activity and thermal stability of tyrosine phenol-lyase by directed evolution.
|
| |
FEBS J,
276,
6187-6194.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

