 |
PDBsum entry 2vg3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Rv2361 with citronellyl pyrophosphate
|
|
Structure:
|
 |
Undecaprenyl pyrophosphate synthetase. Chain: a, b, c, d. Fragment: residues 13-296. Synonym: upp synthetase, undecaprenyl diphosphate synthase, di-trans poly-cis-decaprenylcistransferase, uds, rv2361. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 1773. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.204
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
J.H.Naismith,W.Wang,C.Dong
|
Key ref:

|
 |
W.Wang
et al.
(2008).
The structural basis of chain length control in Rv1086.
J Mol Biol,
381,
129-140.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-Nov-07
|
Release date:
|
06-May-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WFF7
(DPDS_MYCTU) -
Decaprenyl diphosphate synthase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
296 a.a.
284 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.86
- trans,polycis-decaprenyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2Z,6E)-farnesyl diphosphate + 7 isopentenyl diphosphate = (2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E)-decaprenyl diphosphate + 7 diphosphate
|
 |
 |
 |
 |
 |
(2Z,6E)-farnesyl diphosphate
Bound ligand (Het Group name = )
matches with 79.17% similarity
|
+
|
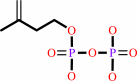 7
×
isopentenyl diphosphate
7
×
isopentenyl diphosphate
|
=
|
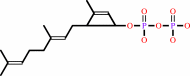 (2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E)-decaprenyl diphosphate
(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E)-decaprenyl diphosphate
|
+
|
 7
×
diphosphate
7
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.87
- ditrans,polycis-polyprenyl diphosphate synthase [(2E,6E)-farnesyl
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
n isopentenyl diphosphate + (2E,6E)-farnesyl diphosphate = a di-trans,poly-cis-polyprenyl diphosphate + n diphosphate
|
 |
 |
 |
 |
 |
n
isopentenyl diphosphate
Bound ligand (Het Group name = )
matches with 79.17% similarity
|
+
|
 7
×
(2E,6E)-farnesyl diphosphate
7
×
(2E,6E)-farnesyl diphosphate
|
=
|
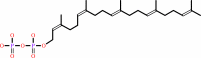 di-trans,poly-cis-polyprenyl diphosphate
di-trans,poly-cis-polyprenyl diphosphate
|
+
|
 n
diphosphate
n
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
381:129-140
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis of chain length control in Rv1086.
|
|
W.Wang,
C.Dong,
M.McNeil,
D.Kaur,
S.Mahapatra,
D.C.Crick,
J.H.Naismith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In Mycobacterium tuberculosis, two related Z-prenyl diphosphate synthases,
E,Z-farnesyl diphosphate synthase (Rv1086) and decaprenyl diphosphate synthase
(Rv2361c), work in series to synthesize decaprenyl phosphate (C(50)) from
isopentenyl diphosphate and E-geranyl diphosphate. Decaprenyl phosphate plays a
central role in the biosynthesis of essential mycobacterial cell wall
components, such as the mycolyl-arabinogalactan-peptidoglycan complex and
lipoarabinomannan; thus, its synthesis has attracted considerable interest as a
potential therapeutic target. Rv1086 is a unique prenyl diphosphate synthase in
that it adds only one isoprene unit to geranyl diphosphate, generating the
15-carbon product (E,Z-farnesyl diphosphate). Rv2361c then adds a further seven
isoprene units to E,Z-farnesyl diphosphate in a processive manner to generate
the 50-carbon prenyl diphosphate, which is then dephosphorylated to generate a
carrier for activated sugars. The molecular basis for chain-length
discrimination by Rv1086 during synthesis is unknown. We also report the
structure of apo Rv1086 with citronellyl diphosphate bound and with the product
mimic E,E-farnesyl diphosphate bound. We report the structures of Rv2361c in the
apo form, with isopentenyl diphosphate bound and with a substrate analogue,
citronellyl diphosphate. The structures confirm the enzymes are very closely
related. Detailed comparison reveals structural differences that account for
chain-length control in Rv1086. We have tested this hypothesis and have
identified a double mutant of Rv1086 that makes a range of longer lipid chains.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Rv1086 and Rv2361c with the reactions they
catalyze. (a) Multiple sequence alignment of open reading frames
Rv1086 and Rv2361c from M. tuberculosis H37Rv with UDPS from M.
luteus (SWISS-PROT O82827) and E. coli (SWISS-PROT Q47675). The
alignment was generated using the Multalin interface. The
N-terminal extension of 2361c is evident. (b) The molecules
discussed in the paper. The product of Rv1086, EZ-FPP, is the
preferred substrate for Rv2361c; we employed EE-FPP in our
crystallographic studies to mimic this molecule. CITPP is an
inhibitor of both enzymes and is a mimic of prenyl diphosphates.
The phosphates are labeled as α and β to distinguish them. (c)
Schematic diagram of the reaction catalyzed by Z-prenyl
synthases. In Rv1086, the reaction is not processive as it stops
after one cycle; in Rv2361c, the reaction continues for a
further seven cycles to make a 50-carbon product.
|
 |
Figure 3.
Fig. 3. The crucial role of L84 in limiting chain length in
Rv1086. (a) Superposition of the active site of EE-FPP Rv1086
complex (protein carbon atoms are white; all other atoms are
colored as in Fig. 2b) and Rv2361c (protein carbon atoms are
colored salmon). The double mutant Rv1086[L84A, L85F] can be
expressed in a soluble (folded) form. L85 in Rv1086 points away
from product. We suggest the L85F mutation role assists only in
folding protein. (b) Space-filling view of the side chains that
limit the growing polymer in Rv1086. EE-FPP is shown as yellow
space-filling spheres. It can be seen that the side chain of L84
covers the polymer. Carbon atoms are in grey for the protein and
yellow for the ligand.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
381,
129-140)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Sato,
K.Takizawa,
Y.Orito,
H.Kudo,
and
T.Hoshino
(2010).
Insight into C35 terpene biosyntheses by nonpathogenic Mycobacterium Species: functional analyses of three Z-prenyltransferases and identification of dehydroheptaprenylcyclines.
|
| |
Chembiochem,
11,
1874-1881.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

