 |
PDBsum entry 2v5c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Family 84 glycoside hydrolase from clostridium perfringens, 2.1 angstrom structure
|
|
Structure:
|
 |
O-glcnacase nagj. Chain: a, b. Fragment: catalytic module, residues 31-624. Synonym: beta-hexosaminidase, hexosaminidase b, gh84, n-acetyl-beta- glucosaminidase, beta-n-acetylhexosaminidase, family 84 glycoside hydrolase. Engineered: yes
|
|
Source:
|
 |
Clostridium perfringens. Organism_taxid: 1502. Atcc: 13124. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.198
|
R-free:
|
0.255
|
|
|
Authors:
|
 |
E.Ficko-Blean,K.J.Gregg,J.J.Adams,J.H.Hehemann,S.J.Smith,M.Czjzek, A.B.Boraston
|
Key ref:

|
 |
E.Ficko-Blean
et al.
(2009).
Portrait of an enzyme, a complete structural analysis of a multimodular {beta}-N-acetylglucosaminidase from Clostridium perfringens.
J Biol Chem,
284,
9876-9884.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Oct-08
|
Release date:
|
27-Jan-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q0TR53
(OGA_CLOP1) -
O-GlcNAcase NagJ from Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1001 a.a.
584 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.169
- protein O-GlcNAcase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
3-O-(N-acetyl-beta-D-glucosaminyl)-L-seryl-[protein] + H2O = N-acetyl-D-glucosamine + L-seryl-[protein]
|
|
2.
|
3-O-(N-acetyl-beta-D-glucosaminyl)-L-threonyl-[protein] + H2O = L-threonyl-[protein] + N-acetyl-D-glucosamine
|
|
 |
 |
 |
 |
 |
3-O-(N-acetyl-beta-D-glucosaminyl)-L-seryl-[protein]
|
+
|
H2O
|
=
|
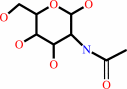 N-acetyl-D-glucosamine
N-acetyl-D-glucosamine
|
+
|
L-seryl-[protein]
|
|
 |
 |
 |
 |
 |
3-O-(N-acetyl-beta-D-glucosaminyl)-L-threonyl-[protein]
|
+
|
H2O
|
=
|
L-threonyl-[protein]
|
+
|
 N-acetyl-D-glucosamine
N-acetyl-D-glucosamine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
284:9876-9884
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Portrait of an enzyme, a complete structural analysis of a multimodular {beta}-N-acetylglucosaminidase from Clostridium perfringens.
|
|
E.Ficko-Blean,
K.J.Gregg,
J.J.Adams,
J.H.Hehemann,
M.Czjzek,
S.P.Smith,
A.B.Boraston.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Common features of the extracellular carbohydrate-active virulence factors
involved in host-pathogen interactions are their large sizes and modular
complexities. This has made them recalcitrant to structural analysis, and
therefore our understanding of the significance of modularity in these important
proteins is lagging. Clostridium perfringens is a prevalent human pathogen that
harbors a wide array of large, extracellular carbohydrate-active enzymes and is
an excellent and relevant model system to approach this problem. Here we
describe the complete structure of C. perfringens GH84C (NagJ), a 1001-amino
acid multimodular homolog of the C. perfringens micro-toxin, which was
determined using a combination of small angle x-ray scattering and x-ray
crystallography. The resulting structure reveals unprecedented insight into how
catalysis, carbohydrate-specific adherence, and the formation of molecular
complexes with other enzymes via an ultra-tight protein-protein interaction are
spatially coordinated in an enzyme involved in a host-pathogen interaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Structures of GH84C catalytic module and GH84C-CBM32 as
determined using x-ray crystallography and SAXS. A and B show
the crystal structures of GH84C catalytic module and
GH84C-CBM32, respectively, in a ribbon representation. The arrow
in B shows the C terminus of the CBM. C shows the
GASBOR-generated SAXS envelope of GH84C-CBM32, whereas D shows
the modules of GH84C-CBM32 manually fit into the SAXS envelope.
E shows the model in D without the SAXS form. F shows the
unmodified x-ray crystal structure, shown in B, fit into the
SAXS-generated envelope. All of the structures are shown from
identical orientations. The N-terminal domain is pictured in
light blue, the catalytic TIM barrel is in orange, the helical
bundle is in pale green, and the CBM in red.
|
 |
Figure 4.
Structural features of the Coh-FN3 modular pair. A shows a
ribbon representation of the 1.8-Å crystal structure of
Coh-FN3. The Coh module is depicted in blue, and FN3 is shown in
black. B shows the surface representation of Coh-FN3 colored
according to electrostatic potential (red is negative, and blue
is positive). The basic patch of FN3 is circled and expanded to
show a patch of basic residues.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2009,
284,
9876-9884)
copyright 2009.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.C.Dorfmueller,
V.S.Borodkin,
D.E.Blair,
S.Pathak,
I.Navratilova,
and
D.M.van Aalten
(2011).
Substrate and product analogues as human O-GlcNAc transferase inhibitors.
|
| |
Amino Acids,
40,
781-792.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Marquay Markiewicz,
S.Syan,
C.C.Hon,
C.Weber,
D.Faust,
and
N.Guillen
(2011).
A proteomic and cellular analysis of uropods in the pathogen Entamoeba histolytica.
|
| |
PLoS Negl Trop Dis,
5,
e1002.
|
 |
|
|
|
|
 |
M.Voronov-Goldman,
R.Lamed,
I.Noach,
I.Borovok,
M.Kwiat,
S.Rosenheck,
L.J.Shimon,
E.A.Bayer,
and
F.Frolow
(2011).
Noncellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
|
| |
Proteins,
79,
50-60.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.C.Dorfmueller,
and
D.M.van Aalten
(2010).
Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds.
|
| |
FEBS Lett,
584,
694-700.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

