 |
PDBsum entry 2rgo

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2rgo
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.1.1.3.21
- glycerol-3-phosphate oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
sn-glycerol 3-phosphate + O2 = dihydroxyacetone phosphate + H2O2
|
 |
 |
 |
 |
 |
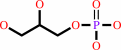 sn-glycerol 3-phosphate
sn-glycerol 3-phosphate
|
+
|
 O2
O2
|
=
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:965-977
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
|
|
T.Colussi,
D.Parsonage,
W.Boles,
T.Matsuoka,
T.C.Mallett,
P.A.Karplus,
A.Claiborne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The FAD-dependent alpha-glycerophosphate oxidase (GlpO) from Enterococcus
casseliflavus and Streptococcus sp. was originally studied as a soluble
flavoprotein oxidase; surprisingly, the GlpO sequence is 30-43% identical to
those of the alpha-glycerophosphate dehydrogenases (GlpDs) from mitochondrial
and bacterial sources. The structure of a deletion mutant of Streptococcus sp.
GlpO (GlpODelta, lacking a 50-residue insert that includes a flexible surface
region) has been determined using multiwavelength anomalous dispersion data and
refined at 2.3 A resolution. Using the GlpODelta structure as a search model, we
have also determined the intact GlpO structure, as refined at 2.4 A resolution.
The first two domains of the GlpO fold are most closely related to those of the
flavoprotein glycine oxidase, where they function in FAD binding and substrate
binding, respectively; the GlpO C-terminal domain consists of two helix bundles
and is not closely related to any known structure. The flexible surface region
in intact GlpO corresponds to a segment of missing electron density that links
the substrate-binding domain to a betabetaalpha element of the FAD-binding
domain. In accordance with earlier biochemical studies (stabilizations of the
covalent FAD-N5-sulfite adduct and p-quinonoid form of 8-mercapto-FAD),
Ile430-N, Thr431-N, and Thr431-OG are hydrogen bonded to FAD-O2alpha in
GlpODelta, stabilizing the negative charge in these two modified flavins and
facilitating transfer of a hydride to FAD-N5 (from Glp) as well. Active-site
overlays with the glycine oxidase-N-acetylglycine and d-amino acid
oxidase-d-alanine complexes demonstrate that Arg346 of GlpODelta is structurally
equivalent to Arg302 and Arg285, respectively; in both cases, these residues
interact directly with the amino acid substrate or inhibitor carboxylate. The
structural and functional divergence between GlpO and the bacterial and
mitochondrial GlpDs is also discussed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Bizzini,
C.Zhao,
A.Budin-Verneuil,
N.Sauvageot,
J.C.Giard,
Y.Auffray,
and
A.Hartke
(2010).
Glycerol is metabolized in a complex and strain-dependent manner in Enterococcus faecalis.
|
| |
J Bacteriol,
192,
779-785.
|
 |
|
|
|
|
 |
P.R.Rich,
and
A.Maréchal
(2010).
The mitochondrial respiratory chain.
|
| |
Essays Biochem,
47,
1.
|
 |
|
|
|
|
 |
D.F.Bischof,
E.M.Vilei,
and
J.Frey
(2009).
Functional and antigenic properties of GlpO from Mycoplasma mycoides subsp. mycoides SC: characterization of a flavin adenine dinucleotide-binding site deletion mutant.
|
| |
Vet Res,
40,
35.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
|

