 |
PDBsum entry 2rbh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.2.9
- gamma-glutamylcyclotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an alpha-(gamma-L-glutamyl)-L-amino acid = 5-oxo-L-proline + an L-alpha- amino acid
|
 |
 |
 |
 |
 |
alpha-(gamma-L-glutamyl)-L-amino acid
|
=
|
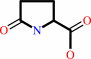 5-oxo-L-proline
5-oxo-L-proline
|
+
|
L-alpha- amino acid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:22031-22042
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle.
|
|
A.J.Oakley,
T.Yamada,
D.Liu,
M.Coggan,
A.G.Clark,
P.G.Board.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The hypothetical protein C7orf24 has been implicated as a cancer marker with a
potential role in cell proliferation. We have identified C7orf24 as
gamma-glutamyl cyclotransferase (GGCT) that catalyzes the formation of
5-oxoproline (pyroglutamic acid) from gamma-glutamyl dipeptides and potentially
plays a significant role in glutathione homeostasis. In the present study we
have identified the first cDNA clones encoding a gamma-glutamyl
cyclotransferase. The GGCT gene is located on chromosome 7p14-15 and consists of
four exons that span 8 kb. The primary sequence is 188 amino acids in length and
is unlike any protein of known function. We crystallized functional recombinant
gamma-glutamyl cyclotransferase and determined its structure at 1.7 A
resolution. The enzyme is a dimer of 20,994-Da subunits. The topology of GGCT is
unrelated to other enzymes associated with cyclotransferase-like activity. The
fold was originally classified as "BtrG-like," a small family that only includes
structures of hypothetical proteins from Mus musculus, Escherichia coli,
Pyrococcus horikoshii, and Arabidopsis thaliana. Since this is the first member
of this family with a defined function, we propose to refer to this structure as
the gamma-glutamyl cyclotransferase fold. We have identified a potential active
site pocket that contains a highly conserved glutamic acid (Glu(98)) and propose
that it acts as a general acid/base in the reaction mechanism. Mutation of
Glu(98) to Ala or Gln completely inactivates the enzyme without altering the
overall fold.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Stereo diagrams of GGCT centered on the putative active site.
Shown are stereo diagrams of the proposed active site regions of
wild type GGCT (A) and E98A (B) and E98Q (C) mutants. Residues
lining the pocket are rendered in stick form. The final 2 mF[o]-
DF[c] electron density maps are rendered in chicken wire form in
orange. All electron density maps are contoured at 1σ. WT, wild
type.
|
 |
Figure 5.
The structure and topology of GGCT. A, schematic diagrams
showing the GGCT dimer. Orthogonal views are shown. Atoms of
Glu^98 are represented as spheres. β-Strands are yellow,
α-helices are purple, and 3[10] helices are green. B, schematic
diagram of the GGCT monomer with secondary structure elements
labeled. C, topology diagram of the GGCT monomer. β-Sheet
interactions are indicated with black boxes. A red box indicates
the interdimer β-sheet interaction. The wrapping of the central
β-sheet to form a barrel is indicated by a faint representation
of strands β2 and β4.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
22031-22042)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
In a subsequent study we have shown that there are at least 3 major protein families with this fold and the human equivalent of the mouse structure shown in figure 9 catalyses a gamma-glutamylamine cyclotransferase reaction that specifically breaks gamma -glutamyl-epsilon -lysine peptide bonds.
Identification and characterization of {gamma}-glutamylamine
cyclotransferase: An enzyme responsible for
{gamma}-glutamyl-{epsilon}-lysine catabolism.
Aaron J. Oakley, Marjorie Coggan, and Philip G. Board
J. Biol. Chem. published 28 January 2010,
10.1074/jbc.M109.082099
http://www.jbc.org/cgi/content/abstract/M109.082099v1?papetoc
Philip Board
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Oda,
S.Makino,
C.Masuda,
T.Yoshiki,
Y.Kitamura,
K.Takata,
D.Yanagisawa,
T.Taniguchi,
and
I.Tooyama
(2009).
The mRNA distribution of C7orf24, a gamma-glutamyl cyclotransferase, in rat tissues.
|
| |
J Histochem Cytochem,
57,
1121-1126.
|
 |
|
|
|
|
 |
L.M.Iyer,
A.M.Burroughs,
and
L.Aravind
(2008).
Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination.
|
| |
Biol Direct,
3,
45.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

