 |
PDBsum entry 2qlp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Bifunctional dctp deaminase:dutpase from mycobacterium tuberculosis, apo form
|
|
Structure:
|
 |
Deoxycytidine triphosphate deaminase. Chain: a, b, c, d, e, f. Synonym: dctp deaminase. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 83332. Strain: h37rv. Gene: dcd. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.47Å
|
R-factor:
|
0.232
|
R-free:
|
0.295
|
|
|
Authors:
|
 |
S.Christophersen,P.Harris,M.Willemoes
|
Key ref:

|
 |
S.S.Helt
et al.
(2008).
Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis.
J Mol Biol,
376,
554-569.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
13-Jul-07
|
Release date:
|
19-Feb-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WP17
(DCDB_MYCTU) -
dCTP deaminase, dUMP-forming from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
190 a.a.
161 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.30
- dCTP deaminase (dUMP-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dCTP + 2 H2O = dUMP + NH4+ + diphosphate
|
 |
 |
 |
 |
 |
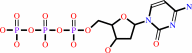 dCTP
dCTP
|
+
|
2
×
H2O
|
=
|
 dUMP
dUMP
|
+
|
NH4(+)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
376:554-569
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis.
|
|
S.S.Helt,
M.Thymark,
P.Harris,
C.Aagaard,
J.Dietrich,
S.Larsen,
M.Willemoes.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Recombinant deoxycytidine triphosphate (dCTP) deaminase from Mycobacterium
tuberculosis was produced in Escherichia coli and purified. The enzyme proved to
be a bifunctional dCTP deaminase:deoxyuridine triphosphatase. As such, the M.
tuberculosis enzyme is the second bifunctional enzyme to be characterised and
provides evidence for bifunctionality of dCTP deaminase occurring outside the
Archaea kingdom. A steady-state kinetic analysis revealed that the affinity for
dCTP and deoxyuridine triphosphate as substrates for the synthesis of
deoxyuridine monophosphate were very similar, a result that contrasts that
obtained previously for the archaean Methanocaldococcus jannaschii enzyme, which
showed approximately 10-fold lower affinity for deoxyuridine triphosphate than
for dCTP. The crystal structures of the enzyme in complex with the inhibitor,
thymidine triphosphate, and the apo form have been solved. Comparison of the two
shows that upon binding of thymidine triphosphate, the disordered C-terminal
arranges as a lid covering the active site, and the enzyme adapts an inactive
conformation as a result of structural changes in the active site. In the
inactive conformation dephosphorylation cannot take place due to the absence of
a water molecule otherwise hydrogen-bonded to O2 of the alpha-phosphate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Structure of M. tuberculosis dCTP deaminase:dUTPase.
Ribbon view of the trimer generated of three A subunits viewed
along the 3-fold axis (a) and perpendicular to the 3-fold axis
(b). The three subunits are shown in cyan, pink and green,
respectively. The nucleotides in the three active sites are
shown in yellow ball-and-stick representation. N- and C-termini
are presented in blue and red, respectively. (c) Schematic view
of the hydrogen-bonding network surrounding dTTP in the
structure of the M. tuberculosis dCTP deaminase:dUTPase–dTTP
complex. The active site is composed of residues from two
subunits indicated by an asterisk (e.g., Ala*). The dotted lines
represent hydrogen bonds between atoms. (d) Electron density
maps presenting the nucleotide in subunit A. The 2F[obs] −
F[calc] map contoured at 1 σ is represented in blue and the
F[obs] − F[calc] map contoured at 3 σ is represented in green.
|
 |
Figure 8.
Fig. 8. dTTP inhibition of the dUTPase reaction in dCTP
deaminase:dUTPase. Close-up of the active site (ribbon view)
from superposition of the apo form of M. tuberculosis dCTP
deaminase:dUTPase (green) and the dTTP complex (pink) with the
M. tuberculosis dUTPase–α,β-imido dUTP complex (orange, PDB
ID 1SIX^24). From all three structures the side chains
corresponding to Ser102, Asp119 and Gln148 are shown as well as
Ala115 from the bifunctional enzyme. Nucleotides are presented
in ball-and-stick representation and in standard atom colours
except carbon, which is coloured according to the matching
enzyme. Important water molecules are also shown in colours
corresponding to the colour of the matching enzyme. Magnesium
ions are grey. The active site is shown from two directions (a
and b). A continuous arrow points to the nucleophilic water
molecule, while a dotted arrow points to the water molecule that
is hydrogen-bonded to O2 of the α-phosphate in the trimeric
dUTPases. See the text for details. The panels were prepared
with PyMol (DeLano Scientific).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
376,
554-569)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.G.Vértessy,
and
J.Tóth
(2009).
Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases.
|
| |
Acc Chem Res,
42,
97.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

