 |
PDBsum entry 2qjh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
M. Jannaschii adh synthase covalently bound to dihydroxyacetone phosphate
|
|
Structure:
|
 |
Putative aldolase mj0400. Chain: a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t. Engineered: yes
|
|
Source:
|
 |
Methanocaldococcus jannaschii. Organism_taxid: 2190. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.203
|
R-free:
|
0.244
|
|
|
Authors:
|
 |
S.E.Ealick,M.Morar
|
|
Key ref:
|
 |
M.Morar
et al.
(2007).
Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry,
46,
10562-10571.
PubMed id:
|
 |
|
Date:
|
 |
|
07-Jul-07
|
Release date:
|
30-Oct-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q57843
(ADHS_METJA) -
2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase from Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
273 a.a.
264 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.10
- 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-deoxy-D-threo-hexo-2,5-diulose 6-phosphate + L-aspartate 4-semialdehyde = 2,3-dioxopropyl phosphate + 2-amino-2,3,7-trideoxy-D-lyxo-hept-6- ulosonate
|
 |
 |
 |
 |
 |
1-deoxy-D-threo-hexo-2,5-diulose 6-phosphate
|
+
|
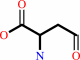 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
=
|
2,3-dioxopropyl phosphate
|
+
|
2-amino-2,3,7-trideoxy-D-lyxo-hept-6- ulosonate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
46:10562-10571
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
|
|
M.Morar,
R.H.White,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Genes responsible for the generation of 3-dehydroquinate (DHQ), an early
metabolite in the established shikimic pathway of aromatic amino acid
biosynthesis, are absent in most euryarchaeotes. Alternative gene products,
Mj0400 and Mj1249, have been identified in Methanocaldococcus jannaschii as the
enzymes involved in the synthesis of DHQ.
2-Amino-3,7-dideoxy-d-threo-hept-6-ulosonic acid (ADH) synthase, the product of
the Mj0400 gene, catalyzes a transaldol reaction between 6-deoxy-5-ketofructose
1-phosphate and l-aspartate semialdehyde to yield ADH. Dehydroquinate synthase
II, the product of the Mj1249 gene, then catalyzes deamination and cyclization
of ADH, resulting in DHQ, which is fed into the canonical pathway. Three crystal
structures of ADH synthase were determined in this work: a complex with a
substrate analogue, fructose 1,6-bisphosphate, a complex with dihydroxyacetone
phosphate (DHAP), thought to be a product of fructose 1-phosphate cleavage, and
a native structure containing copurified ligands, modeled as DHAP and glycerol.
On the basis of the structural analysis and comparison of the enzyme with
related aldolases, ADH synthase is classified as a new member of the class I
aldolase superfamily. The description of the active site allows for the
identification and characterization of possible catalytic residues, Lys184,
which is responsible for formation of the Schiff base intermediate, and Asp33
and Tyr153, which are candidates for the general acid/base catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Samland,
and
G.A.Sprenger
(2009).
Transaldolase: from biochemistry to human disease.
|
| |
Int J Biochem Cell Biol,
41,
1482-1494.
|
 |
|
|
|
|
 |
Z.Diaz,
K.B.Xavier,
and
S.T.Miller
(2009).
The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
|
| |
PLoS One,
4,
e6820.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Samland,
M.Wang,
and
G.A.Sprenger
(2008).
MJ0400 from Methanocaldococcus jannaschii exhibits fructose-1,6-bisphosphate aldolase activity.
|
| |
FEMS Microbiol Lett,
281,
36-41.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

