 |
PDBsum entry 2qdk

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
X-ray structure of the unliganded uridine phosphorylase from salmonella typhimurium at 1.62a resolution
|
|
Structure:
|
 |
Uridine phosphorylase. Chain: a, b, c, d, e, f. Synonym: urdpase, upase. Engineered: yes
|
|
Source:
|
 |
Salmonella typhimurium. Strain: lt2. Gene: udp. Expressed in: escherichia coli.
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
V.I.Timofeev,B.P.Pavlyuk,A.A.Lashkov,A.G.Gabdoulkhakov,A.M.Mikhailov
|
|
Key ref:
|
 |
V.I.Timofeev
et al.
X-Ray structure of the unliganded uridine phosphorylase from salmonella typhimurium at 1.62a resolution.
To be published,
.
|
 |
|
Date:
|
 |
|
21-Jun-07
|
Release date:
|
01-Jul-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.2.4.2.3
- uridine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uridine + phosphate = alpha-D-ribose 1-phosphate + uracil
|
 |
 |
 |
 |
 |
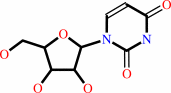 uridine
uridine
|
+
|
 phosphate
phosphate
|
=
|
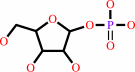 alpha-D-ribose 1-phosphate
alpha-D-ribose 1-phosphate
|
+
|
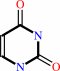 uracil
uracil
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

