 |
PDBsum entry 2q5h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of apo-wildtype glycyl-tRNA synthetase
|
|
Structure:
|
 |
Glycyl-tRNA synthetase. Chain: a. Fragment: residues 55-739. Synonym: glycine-tRNA ligase, glyrs. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: gars. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.207
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
M.Z.Cader,J.Ren,P.A.James,L.E.Bird,K.Talbot,D.K.Stammers,Oxford Protein Production Facility (Oppf)
|
|
Key ref:
|
 |
M.Z.Cader
et al.
(2007).
Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett,
581,
2959-2964.
PubMed id:
|
 |
|
Date:
|
 |
|
01-Jun-07
|
Release date:
|
19-Jun-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P41250
(GARS_HUMAN) -
Glycine--tRNA ligase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
739 a.a.
528 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.1.1.14
- glycine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Gly) + glycine + ATP = glycyl-tRNA(Gly) + AMP + diphosphate
|
 |
 |
 |
 |
 |
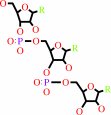 tRNA(Gly)
tRNA(Gly)
|
+
|
 glycine
glycine
|
+
|
 ATP
ATP
|
=
|
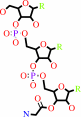 glycyl-tRNA(Gly)
glycyl-tRNA(Gly)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Febs Lett
581:2959-2964
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
|
|
M.Z.Cader,
J.Ren,
P.A.James,
L.E.Bird,
K.Talbot,
D.K.Stammers.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dominant mutations in the ubiquitous enzyme glycyl-tRNA synthetase (GlyRS),
including S581L, lead to motor nerve degeneration. We have determined crystal
structures of wildtype and S581L-mutant human GlyRS. The S581L mutation is
approximately 50A from the active site, and yet gives reduced aminoacylation
activity. The overall structures of wildtype and S581L-GlyRS, including the
active site, are very similar. However, residues 567-575 of the
anticodon-binding domain shift position and in turn could indirectly affect
glycine binding via the tRNA or alternatively inhibit conformational changes.
Reduced enzyme activity may underlie neuronal degeneration, although a
dominant-negative effect is more likely in this autosomal dominant disorder.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.W.Motley,
K.Talbot,
and
K.H.Fischbeck
(2010).
GARS axonopathy: not every neuron's cup of tRNA.
|
| |
Trends Neurosci,
33,
59-66.
|
 |
|
|
|
|
 |
F.Achilli,
V.Bros-Facer,
H.P.Williams,
G.T.Banks,
M.AlQatari,
R.Chia,
V.Tucci,
M.Groves,
C.D.Nickols,
K.L.Seburn,
R.Kendall,
M.Z.Cader,
K.Talbot,
J.van Minnen,
R.W.Burgess,
S.Brandner,
J.E.Martin,
M.Koltzenburg,
L.Greensmith,
P.M.Nolan,
and
E.M.Fisher
(2009).
An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy.
|
| |
Dis Model Mech,
2,
359-373.
|
 |
|
|
|
|
 |
R.T.Guo,
Y.E.Chong,
M.Guo,
and
X.L.Yang
(2009).
Crystal structures and biochemical analyses suggest a unique mechanism and role for human glycyl-tRNA synthetase in Ap4A homeostasis.
|
| |
J Biol Chem,
284,
28968-28976.
|
 |
|
|
|
|
 |
A.Antonellis,
and
E.D.Green
(2008).
The role of aminoacyl-tRNA synthetases in genetic diseases.
|
| |
Annu Rev Genomics Hum Genet,
9,
87.
|
 |
|
|
|
|
 |
S.G.Park,
P.Schimmel,
and
S.Kim
(2008).
Aminoacyl tRNA synthetases and their connections to disease.
|
| |
Proc Natl Acad Sci U S A,
105,
11043-11049.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

