 |
PDBsum entry 2q1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.141
- 2-dehydro-3-deoxy-D-arabinonate dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-dehydro-3-deoxy-D-arabinonate = 2,5-dioxopentanoate + H2O
|
 |
 |
 |
 |
 |
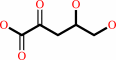 2-dehydro-3-deoxy-D-arabinonate
2-dehydro-3-deoxy-D-arabinonate
|
=
|
2,5-dioxopentanoate
Bound ligand (Het Group name = )
matches with 77.78% similarity
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
379:357-371
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
|
|
S.J.Brouns,
T.R.Barends,
P.Worm,
J.Akerboom,
A.P.Turnbull,
L.Salmon,
J.van der Oost.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The archaeon Sulfolobus solfataricus converts d-arabinose to 2-oxoglutarate by
an enzyme set consisting of two dehydrogenases and two dehydratases. The third
step of the pathway is catalyzed by a novel 2-keto-3-deoxy-D-arabinonate
dehydratase (KdaD). In this study, the crystal structure of the enzyme has been
solved to 2.1 A resolution. The enzyme forms an oval-shaped ring of four
subunits, each consisting of an N-terminal domain with a four-stranded
beta-sheet flanked by two alpha-helices, and a C-terminal catalytic domain with
a fumarylacetoacetate hydrolase (FAH) fold. Crystal structures of complexes of
the enzyme with magnesium or calcium ions and either a substrate analog
2-oxobutyrate, or the aldehyde enzyme product 2,5-dioxopentanoate revealed that
the divalent metal ion in the active site is coordinated octahedrally by three
conserved carboxylate residues, a water molecule, and both the carboxylate and
the oxo groups of the substrate molecule. An enzymatic mechanism for
base-catalyzed dehydration is proposed on the basis of the binding mode of the
substrate to the metal ion, which suggests that the enzyme enhances the acidity
of the protons alpha to the carbonyl group, facilitating their abstraction by
glutamate 114. A comprehensive structural comparison of members of the FAH
superfamily is presented and their evolution is discussed, providing a basis for
functional investigations of this largely unexplored protein superfamily.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. (a) Topology diagram showing the connectivity of
secondary structure elements and domain organization of the S.
solfataricus KdaD monomer. The N terminus of the polypeptide
chain (N, in blue) follows a color gradient towards the C
terminus (C, in red). (b and c) Ribbon diagrams at two viewing
angles of the KdaD monomer with Mg^2+ and 2-oxobutyrate bound.
|
 |
Figure 4.
Fig. 4. Stereo diagrams are shown of the KdaD active site
with Mg^2+ (green spheres), water molecules (red spheres) and
(a) 2,5-dioxopentanoate or (b) 2-oxobutyrate bound. To exclude
any model bias, simulated annealing F[o]–DF[c] omit electron
density maps for the ligand and the metal ion are displayed at
the 3σ contour level. (c) Stereo image of a model of the
Michaelis complex of KdaD with its substrate
2-keto-3-deoxy-d-arabinonate (D-KDA) bound. Hydrogen atoms for
D-KDA are shown as well as the carbon atom numbers.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
379,
357-371)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Watanabe,
and
K.Makino
(2009).
Novel modified version of nonphosphorylated sugar metabolism--an alternative L-rhamnose pathway of Sphingomonas sp.
|
| |
FEBS J,
276,
1554-1567.
|
 |
|
|
|
|
 |
U.Johnsen,
M.Dambeck,
H.Zaiss,
T.Fuhrer,
J.Soppa,
U.Sauer,
and
P.Schönheit
(2009).
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
| |
J Biol Chem,
284,
27290-27303.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

