 |
PDBsum entry 2q0s

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structure of the inhibitor bound form of m. Smegmatis aryl esterase
|
|
Structure:
|
 |
Aryl esterase. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Mycobacterium smegmatis. Organism_taxid: 1772. Strain: atcc19686. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.135
|
R-free:
|
0.160
|
|
|
Authors:
|
 |
I.I.Mathews,M.Soltis,M.Saldajeno,G.Ganshaw,R.Sala,W.Weyler, M.A.Cervin,G.Whited,R.Bott
|
|
Key ref:
|
 |
I.Mathews
et al.
(2007).
Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions.
Biochemistry,
46,
8969-8979.
PubMed id:
|
 |
|
Date:
|
 |
|
22-May-07
|
Release date:
|
11-Dec-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0R5U7
(A0R5U7_MYCS2) -
Lipolytic enzyme, G-D-S-L from Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
216 a.a.
215 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.2
- arylesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phenyl acetate + H2O = a phenol + acetate + H+
|
 |
 |
 |
 |
 |
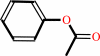 phenyl acetate
phenyl acetate
|
+
|
H2O
|
=
|
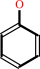 phenol
phenol
|
+
|
 acetate
acetate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
46:8969-8979
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions.
|
|
I.Mathews,
M.Soltis,
M.Saldajeno,
G.Ganshaw,
R.Sala,
W.Weyler,
M.A.Cervin,
G.Whited,
R.Bott.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The unusual architecture of the enzyme (MsAcT) isolated from Mycobacterium
smegmatis forms the mechanistic basis for favoring alcoholysis over hydrolysis
in water. Unlike hydrolases that perform alcoholysis only under anhydrous
conditions, MsAcT demonstrates alcoholysis in substantially aqueous media and,
in the presence of hydrogen peroxide, has a perhydrolysis:hydrolysis ratio
50-fold greater than that of the best lipase tested. The crystal structures of
the apoenzyme and an inhibitor-bound form have been determined to 1.5 A
resolution. MsAcT is an octamer in the asymmetric unit and forms a tightly
associated aggregate in solution. Relative to other structurally similar
monomers, MsAcT contains several insertions that contribute to the
oligomerization and greatly restrict the shape of the active site, thereby
limiting its accessibility. These properties create an environment by which
MsAcT can catalyze transesterification reactions in an aqueous medium and
suggests how a serine hydrolase can be engineered to be an efficient
acyltransferase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Ma,
Q.Lu,
Y.Yuan,
H.Ge,
K.Li,
W.Zhao,
Y.Gao,
L.Niu,
and
M.Teng
(2011).
Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity.
|
| |
Proteins,
79,
662-668.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Duncan,
Q.Jing,
A.Katona,
R.J.Kazlauskas,
J.Schilling,
U.Tschirner,
and
W.W.Aldajani
(2010).
Increased saccharification yields from aspen biomass upon treatment with enzymatically generated peracetic acid.
|
| |
Appl Biochem Biotechnol,
160,
1637-1652.
|
 |
|
|
|
|
 |
S.Santini,
J.M.Crowet,
A.Thomas,
M.Paquot,
M.Vandenbol,
P.Thonart,
J.P.Wathelet,
C.Blecker,
G.Lognay,
R.Brasseur,
L.Lins,
and
B.Charloteaux
(2009).
Study of Thermomyces lanuginosa lipase in the presence of tributyrylglycerol and water.
|
| |
Biophys J,
96,
4814-4825.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

