 |
PDBsum entry 2pw6

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
2pw6
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, unknown function
|
 |
|
Title:
|
 |
Crystal structure of uncharacterized protein jw3007 from escherichia coli k12
|
|
Structure:
|
 |
Uncharacterized protein ygid. Chain: a. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 83333. Strain: k12. Gene: ygid, b3039, jw3007. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.27Å
|
R-factor:
|
0.236
|
R-free:
|
0.267
|
|
|
Authors:
|
 |
M.G.Newton,Y.Takagi,A.Ebihara,A.Shinkai,S.Kuramitsu,S.Yokayama,Y.Li, L.Chen,J.Zhu,J.Ruble,Z.J.Liu,J.P.Rose,B.-C.Wang,Southeast Collaboratory For Structural Genomics (Secsg),Riken Structural Genomics/proteomics Initiative (Rsgi)
|
|
Key ref:
|
 |
M.G.Newton
et al.
Crystal structure of uncharacterized protein jw3007 from escherichia coli k12..
To be published,
.
|
 |
|
Date:
|
 |
|
10-May-07
|
Release date:
|
12-Jun-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P24197
(YGID_ECOLI) -
4,5-DOPA dioxygenase extradiol from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
271 a.a.
244 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.29
- stizolobate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-dopa + O2 = 4-(L-alanin-3-yl)-2-hydroxy-cis,cis-muconate 6-semialdehyde + H+
|
 |
 |
 |
 |
 |
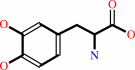 L-dopa
L-dopa
|
+
|
 O2
O2
|
=
|
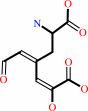 4-(L-alanin-3-yl)-2-hydroxy-cis,cis-muconate 6-semialdehyde
4-(L-alanin-3-yl)-2-hydroxy-cis,cis-muconate 6-semialdehyde
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

