 |
PDBsum entry 2prl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2prl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.5.2
- dihydroorotate dehydrogenase (quinone).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-dihydroorotate + a quinone = orotate + a quinol
|
 |
 |
 |
 |
 |
(S)-dihydroorotate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 quinone
quinone
|
=
|
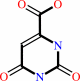 orotate
orotate
|
+
|
quinol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:8929-8936
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites.
|
|
B.Walse,
V.T.Dufe,
B.Svensson,
I.Fritzson,
L.Dahlberg,
A.Khairoullina,
U.Wellmar,
S.Al-Karadaghi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Inhibitors of dihydroorotate dehydrogenase (DHODH) have been suggested for the
treatment of rheumatoid arthritis, psoriasis, autoimmune diseases, Plasmodium,
and bacterial and fungal infections. Here we present the structures of
N-terminally truncated (residues Met30-Arg396) DHODH in complex with two
inhibitors: a brequinar analogue (6) and a novel inhibitor (a fenamic acid
derivative) (7), as well as the first structure of the enzyme to be
characterized without any bound inhibitor. It is shown that 7 uses the
"standard" brequinar binding mode and, in addition, interacts with Tyr356, a
residue conserved in most class 2 DHODH proteins. Compared to the inhibitor-free
structure, some of the amino acid side chains in the tunnel in which brequinar
binds and which was suggested to be the binding site of ubiquinone undergo
changes in conformation upon inhibitor binding. Using our data, the loop regions
of residues Leu68-Arg72 and Asn212-Leu224, which were disordered in previously
studied human DHODH structures, could be built into the electron density. The
first of these loops, which is located at the entrance to the inhibitor-binding
pocket, shows different conformations in the three structures, suggesting that
it may interfere with inhibitor/cofactor binding. The second loop has been
suggested to control the access of dihydroorotate to the active site of the
enzyme and may be an important player in the enzymatic reaction. These
observations provide new insights into the dynamic features of the DHODH
reaction and suggest new approaches to the design of inhibitors against DHODH.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Lenaz,
and
M.L.Genova
(2010).
Structure and organization of mitochondrial respiratory complexes: a new understanding of an old subject.
|
| |
Antioxid Redox Signal,
12,
961.
|
 |
|
|
|
|
 |
I.Fritzson,
B.Svensson,
S.Al-Karadaghi,
B.Walse,
U.Wellmar,
U.J.Nilsson,
D.da Graça Thrige,
and
S.Jönsson
(2010).
Inhibition of human DHODH by 4-hydroxycoumarins, fenamic acids, and N-(alkylcarbonyl)anthranilic acids identified by structure-guided fragment selection.
|
| |
ChemMedChem,
5,
608-617.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.A.Phillips,
and
P.K.Rathod
(2010).
Plasmodium dihydroorotate dehydrogenase: a promising target for novel anti-malarial chemotherapy.
|
| |
Infect Disord Drug Targets,
10,
226-239.
|
 |
|
|
|
|
 |
P.R.Rich,
and
A.Maréchal
(2010).
The mitochondrial respiratory chain.
|
| |
Essays Biochem,
47,
1.
|
 |
|
|
|
|
 |
R.L.Kow,
J.R.Whicher,
C.A.McDonald,
B.A.Palfey,
and
R.L.Fagan
(2009).
Disruption of the proton relay network in the class 2 dihydroorotate dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
48,
9801-9809.
|
 |
|
|
|
|
 |
X.Deng,
R.Gujjar,
F.El Mazouni,
W.Kaminsky,
N.A.Malmquist,
E.J.Goldsmith,
P.K.Rathod,
and
M.A.Phillips
(2009).
Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
|
| |
J Biol Chem,
284,
26999-27009.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

