 |
PDBsum entry 2plq

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.4
- amidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a monocarboxylic acid amide + H2O = a monocarboxylate + NH4+
|
 |
 |
 |
 |
 |
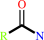 monocarboxylic acid amide
monocarboxylic acid amide
|
+
|
H2O
|
=
|
 monocarboxylate
monocarboxylate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
62:1174-1178
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The quaternary structure of the amidase from Geobacillus pallidus RAPc8 is revealed by its crystal packing.
|
|
V.B.Agarkar,
S.W.Kimani,
D.A.Cowan,
M.F.Sayed,
B.T.Sewell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The amidase from Geobacillus pallidus RAPc8, a moderate thermophile, is a member
of the nitrilase enzyme superfamily. It converts amides to the corresponding
acids and ammonia and has application as an industrial catalyst. RAPc8 amidase
has been cloned and functionally expressed in Escherichia coli and has been
purified by heat treatment and a number of chromatographic steps. The enzyme was
crystallized using the hanging-drop vapour-diffusion method. Crystals produced
in the presence of 1.2 M sodium citrate, 400 mM NaCl, 100 mM sodium acetate pH
5.6 were selected for X-ray diffraction studies. A data set having acceptable
statistics to 1.96 A resolution was collected under cryoconditions using an
in-house X-ray source. The space group was determined to be primitive cubic
P4(2)32, with unit-cell parameter a = 130.49 (+/-0.05) A. The structure was
solved by molecular replacement using the backbone of the hypothetical protein
PH0642 from Pyrococcus horikoshii (PDB code 1j31) with all non-identical side
chains substituted with alanine as a probe. There is one subunit per asymmetric
unit. The subunits are packed as trimers of dimers with D3 point-group symmetry
around the threefold axis in such a way that the dimer interface seen in the
homologues is preserved.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
A crystal of the amidase from G. pallidus RAPc8. Acta
Crystallogr Sect F Struct Biol Cryst Commun. 2006 December 1;
62(Pt 12): 1174–1178. Published online 2006 November 4. doi:
10.1107/S1744309106043855. Copyright [copyright] International
Union of Crystallography 2006
|
 |
Figure 4.
Stereoviews of the suggested solution hexamer viewed down one
of the twofold axes from both directions. The preserved dimer
surface is centred in (a). The interacting surfaces depicted in
the foreground of (b) are based on the model of the putative CN
hydrolase from yeast (PDB code 1f89). 66 C-terminal amino acids
for each subunit are not depicted in this model. Acta
Crystallogr Sect F Struct Biol Cryst Commun. 2006 December 1;
62(Pt 12): 1174–1178. Published online 2006 November 4. doi:
10.1107/S1744309106043855. Copyright [copyright] International
Union of Crystallography 2006
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the IUCr:
Acta Crystallogr Sect F Struct Biol Cryst Commun
(2006,
62,
1174-1178)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Andrade,
A.Karmali,
M.A.Carrondo,
and
C.Frazão
(2007).
Crystallization, diffraction data collection and preliminary crystallographic analysis of hexagonal crystals of Pseudomonas aeruginosa amidase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
214-216.
|
 |
|
|
|
|
 |
R.N.Thuku,
B.W.Weber,
A.Varsani,
and
B.T.Sewell
(2007).
Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form.
|
| |
FEBS J,
274,
2099-2108.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

