 |
PDBsum entry 2pl5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.31
- homoserine O-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homoserine + acetyl-CoA = O-acetyl-L-homoserine + CoA
|
 |
 |
 |
 |
 |
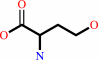 L-homoserine
L-homoserine
|
+
|
acetyl-CoA
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
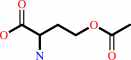 O-acetyl-L-homoserine
O-acetyl-L-homoserine
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem Biophys Res Commun
363:1050-1056
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans.
|
|
M.Wang,
L.Liu,
Y.Wang,
Z.Wei,
P.Zhang,
Y.Li,
X.Jiang,
H.Xu,
W.Gong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Homoserine O-acetyltransferase (HTA, EC 2.3.1.31) initiates methionine
biosynthesis pathway by catalyzing the transfer of acetyl group from acetyl-CoA
to homoserine. This study reports the crystal structure of HTA from Leptospira
interrogans determined at 2.2A resolution using selenomethionyl
single-wavelength anomalous diffraction method. HTA is modular and consists of
two structurally distinct domains--a core alpha/beta domain containing the
catalytic site and a helical bundle called the lid domain. Overall, the
structure fold belongs to alpha/beta hydrolase superfamily with the
characteristic 'catalytic triad' residues in the active site. Detailed structure
analysis showed that the catalytic histidine and serine are both present in two
conformations, which may be involved in the catalytic mechanism for acetyl
transfer.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Bergfeld,
H.Claus,
N.K.Lorenzen,
F.Spielmann,
U.Vogel,
and
M.Mühlenhoff
(2009).
The Polysialic Acid-specific O-Acetyltransferase OatC from Neisseria meningitidis Serogroup C Evolved Apart from Other Bacterial Sialate O-Acetyltransferases.
|
| |
J Biol Chem,
284,
6.
|
 |
|
|
|
|
 |
C.Tölzer,
S.Pal,
H.Watzlawick,
J.Altenbuchner,
and
K.Niefind
(2009).
Crystallization and preliminary crystallographic analysis of cgHle, a homoserine acetyltransferase homologue, from Corynebacterium glutamicum.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
34-38.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

