 |
PDBsum entry 2pid

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of human mitochondrial tyrosyl-tRNA synthetase in complex with an adenylate analog
|
|
Structure:
|
 |
Tyrosyl-tRNA synthetase. Chain: a, b. Fragment: residues 28-375. Synonym: tyrosine-tRNA ligase, tyrrs. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: yars2. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.193
|
R-free:
|
0.244
|
|
|
Authors:
|
 |
L.Bonnefond,M.Frugier,E.Touze,B.Lorber,C.Florentz,R.Giege,C.Sauter, J.Rudinger-Thirion
|
Key ref:

|
 |
L.Bonnefond
et al.
(2007).
Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features.
Structure,
15,
1505-1516.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
13-Apr-07
|
Release date:
|
23-Oct-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9Y2Z4
(SYYM_HUMAN) -
Tyrosine--tRNA ligase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
477 a.a.
317 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.1
- tyrosine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Tyr) + L-tyrosine + ATP = L-tyrosyl-tRNA(Tyr) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
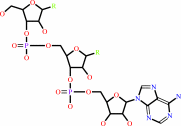 tRNA(Tyr)
tRNA(Tyr)
|
+
|
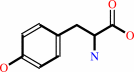 L-tyrosine
L-tyrosine
|
+
|
 ATP
ATP
|
=
|
L-tyrosyl-tRNA(Tyr)
Bound ligand (Het Group name = )
matches with 48.72% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
15:1505-1516
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features.
|
|
L.Bonnefond,
M.Frugier,
E.Touzé,
B.Lorber,
C.Florentz,
R.Giegé,
C.Sauter,
J.Rudinger-Thirion.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We report the structure of a strictly mitochondrial human synthetase, namely
tyrosyl-tRNA synthetase (mt-TyrRS), in complex with an adenylate analog at 2.2 A
resolution. The structure is that of an active enzyme deprived of the C-terminal
S4-like domain and resembles eubacterial TyrRSs with a canonical
tyrosine-binding pocket and adenylate-binding residues typical of class I
synthetases. Two bulges at the enzyme surface, not seen in eubacterial TyrRSs,
correspond to conserved sequences in mt-TyrRSs. The synthetase electrostatic
surface potential differs from that of other TyrRSs, including the human
cytoplasmic homolog and the mitochondrial one from Neurospora crassa. The
homodimeric human mt-TyrRS shows an asymmetry propagating from the dimer
interface toward the two catalytic sites and extremities of each subunit.
Mutagenesis of the catalytic domain reveals functional importance of Ser200 in
line with an involvement of A73 rather than N1-N72 in tyrosine identity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Conformational Asymmetry in Homodimeric Human
mt-TyrRS-ΔS4
Superimposition of the two subunits of
mt-TyrRS-ΔS4 in complex with Tyr-AMS (backbones colored as in
Figure 1A, in heavy and light colors for monomer A and B,
respectively). The regions with largest asymmetries are circled.
Note that the two adenylate analogs (in blue) almost perfectly
superimpose.
|
 |
Figure 7.
Figure 7. Role of Clusters 1 and 2 in tRNA^Tyr Acceptor Arm
Recognition
(A) Superimposition of the cleft formed by the
two helical structures of clusters 1 and 2 (in which binds the
tyrosine acceptor arm of tRNA^Tyr) in the crystallographic
structures of human mt-TyrRS (in brown), B. stearothermophilus
TyrRS (in blue), and T. thermophilus TyrRS in complex with
tRNA^Tyr (in green). Notice the quasiperfect superimposition of
the two clusters in human mt-TyrRS and B. stearothermophilus
TyrRS and the important structural deviations in T. thermophilus
TyrRS. The bar at the bottom of the figure shows the position
where the cleft is largest (d = 9.9, 10.0, and 12.4 Å in
the TyrRSs from human mitochondria, B. stearothermophilus, and
T. thermophilus, respectively, indicating an enlargement of the
cleft in T. thermophilus of  vert,
similar 2.5 Å). The three amino acids that were
mutagenized are indicated. vert,
similar 2.5 Å). The three amino acids that were
mutagenized are indicated.
(B) View of the clusters and
their proximity with the tRNA acceptor branch as seen in the
crystal structure of the T. thermophilus complex (Yaremchuk et
al., 2002).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(2007,
15,
1505-1516)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.G.Riley,
S.Cooper,
P.Hickey,
J.Rudinger-Thirion,
M.McKenzie,
A.Compton,
S.C.Lim,
D.Thorburn,
M.T.Ryan,
R.Giegé,
M.Bahlo,
and
J.Christodoulou
(2010).
Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome.
|
| |
Am J Hum Genet,
87,
52-59.
|
 |
|
|
|
|
 |
R.Giegé,
and
C.Sauter
(2010).
Biocrystallography: past, present, future.
|
| |
HFSP J,
4,
109-121.
|
 |
|
|
|
|
 |
I.A.Vasil'eva,
E.A.Semenova,
and
N.A.Moor
(2009).
Interaction of human phenylalanyl-tRNA synthetase with specific tRNA according to thiophosphate footprinting.
|
| |
Biochemistry (Mosc),
74,
175-185.
|
 |
|
|
|
|
 |
L.Bonnefond,
C.Florentz,
R.Giegé,
and
J.Rudinger-Thirion
(2008).
Decreased aminoacylation in pathology-related mutants of mitochondrial tRNATyr is associated with structural perturbations in tRNA architecture.
|
| |
RNA,
14,
641-648.
|
 |
|
|
|
|
 |
Q.Vicens,
P.J.Paukstelis,
E.Westhof,
A.M.Lambowitz,
and
T.R.Cech
(2008).
Toward predicting self-splicing and protein-facilitated splicing of group I introns.
|
| |
RNA,
14,
2013-2029.
|
 |
|
|
|
|
 |
R.Giegé
(2008).
Toward a more complete view of tRNA biology.
|
| |
Nat Struct Mol Biol,
15,
1007-1014.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

