 |
PDBsum entry 2php

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
2php
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, unknown function
|
 |
|
Title:
|
 |
Crystal structure of thE C-terminal domain of protein mj0236 (y236_metja)
|
|
Structure:
|
 |
Uncharacterized protein mj0236. Chain: a, b, d, e. Fragment: residues 240-420. Engineered: yes
|
|
Source:
|
 |
Methanocaldococcus jannaschii dsm 2661. Organism_taxid: 243232. Strain: dsm 2661, jal-1, jcm 10045, nbrc 100440. Atcc: 43067. Gene: mj0236. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.03Å
|
R-factor:
|
0.216
|
R-free:
|
0.238
|
|
|
Authors:
|
 |
S.Eswaramoorthy,S.K.Burley,S.Swaminathan,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
S.Eswaramoorthy
et al.
Crystal structure of the c-Terminal domain of protein mj0236 (y236_metja).
To be published,
.
|
 |
|
Date:
|
 |
|
11-Apr-07
|
Release date:
|
24-Apr-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q57688
(THIDN_METJA) -
Bifunctional thiamine biosynthesis protein ThiDN from Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
417 a.a.
181 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.3
- thiamine phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
2-[(2R,5Z)-2-carboxy-4-methylthiazol-5(2H)-ylidene]ethyl phosphate + 4-amino-2-methyl-5-(diphosphooxymethyl)pyrimidine + 2 H+ = thiamine phosphate + CO2 + diphosphate
|
|
2.
|
2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate + 4-amino-2-methyl- 5-(diphosphooxymethyl)pyrimidine + 2 H+ = thiamine phosphate + CO2 + diphosphate
|
|
3.
|
4-methyl-5-(2-phosphooxyethyl)-thiazole + 4-amino-2-methyl-5- (diphosphooxymethyl)pyrimidine + H+ = thiamine phosphate + diphosphate
|
|
 |
 |
 |
 |
 |
2-[(2R,5Z)-2-carboxy-4-methylthiazol-5(2H)-ylidene]ethyl phosphate
|
+
|
4-amino-2-methyl-5-(diphosphooxymethyl)pyrimidine
|
+
|
2
×
H(+)
|
=
|
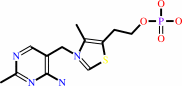 thiamine phosphate
thiamine phosphate
|
+
|
 CO2
CO2
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate
|
+
|
4-amino-2-methyl- 5-(diphosphooxymethyl)pyrimidine
|
+
|
2
×
H(+)
|
=
|
 thiamine phosphate
thiamine phosphate
|
+
|
 CO2
CO2
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
4-methyl-5-(2-phosphooxyethyl)-thiazole
|
+
|
4-amino-2-methyl-5- (diphosphooxymethyl)pyrimidine
|
+
|
H(+)
|
=
|
 thiamine phosphate
thiamine phosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.1.49
- hydroxymethylpyrimidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-amino-5-hydroxymethyl-2-methylpyrimidine + ATP = 4-amino-2-methyl-5- (phosphooxymethyl)pyrimidine + ADP + H+
|
 |
 |
 |
 |
 |
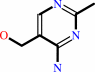 4-amino-5-hydroxymethyl-2-methylpyrimidine
4-amino-5-hydroxymethyl-2-methylpyrimidine
|
+
|
 ATP
ATP
|
=
|
2
×
4-amino-2-methyl-5- (phosphooxymethyl)pyrimidine
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.7.4.7
- phosphooxymethylpyrimidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-amino-2-methyl-5-(phosphooxymethyl)pyrimidine + ATP = 4-amino-2-methyl- 5-(diphosphooxymethyl)pyrimidine + ADP
|
 |
 |
 |
 |
 |
4-amino-2-methyl-5-(phosphooxymethyl)pyrimidine
|
+
|
 ATP
ATP
|
=
|
2
×
4-amino-2-methyl- 5-(diphosphooxymethyl)pyrimidine
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

