 |
PDBsum entry 2pf8

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2pf8
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Complex of aldose reductase with NADP+ and simaltaneously bound competetive inhibitors fidarestat and idd594. Concentration of fidarestat in soaking solution is equal to concentration of idd594.
|
|
Structure:
|
 |
Aldose reductase. Chain: a. Synonym: ar, aldehyde reductase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: akr1b1, aldr1. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
0.85Å
|
R-factor:
|
0.086
|
R-free:
|
0.095
|
|
|
Authors:
|
 |
T.Petrova,I.Hazemann,A.Cousido,A.Mitschler,S.Ginell,A.Joachimiak, A.Podjarny
|
|
Key ref:
|
 |
A.Cousido-Siah
et al.
(2012).
Crystal packing modifies ligand binding affinity: the case of aldose reductase.
Proteins,
80,
2552-2561.
PubMed id:
|
 |
|
Date:
|
 |
|
04-Apr-07
|
Release date:
|
17-Apr-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15121
(ALDR_HUMAN) -
Aldo-keto reductase family 1 member B1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
316 a.a.
316 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.21
- aldose reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an alditol + NAD+ = an aldose + NADH + H+
|
|
2.
|
an alditol + NADP+ = an aldose + NADPH + H+
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldose
aldose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldose
aldose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
all-trans-retinol
Bound ligand (Het Group name = )
matches with 41.38% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
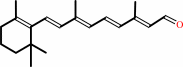 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
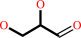 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
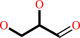 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
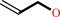 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
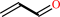 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Proteins
80:2552-2561
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal packing modifies ligand binding affinity: the case of aldose reductase.
|
|
A.Cousido-Siah,
T.Petrova,
I.Hazemann,
A.Mitschler,
F.X.Ruiz,
E.Howard,
S.Ginell,
C.Atmanene,
A.Van Dorsselaer,
S.Sanglier-Cianférani,
A.Joachimiak,
A.Podjarny.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

