 |
PDBsum entry 2pdp

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2pdp
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.21
- aldose reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an alditol + NAD+ = an aldose + NADH + H+
|
|
2.
|
an alditol + NADP+ = an aldose + NADPH + H+
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldose
aldose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldose
aldose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
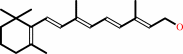 all-trans-retinol
all-trans-retinol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
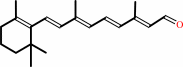 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
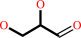 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
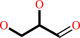 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
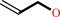 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
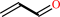 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
379:991-1016
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
|
|
H.Steuber,
A.Heine,
A.Podjarny,
G.Klebe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Inhibition of human aldose reductase (ALR2) evolved as a promising therapeutic
concept to prevent late complications of diabetes. As well as appropriate
affinity and bioavailability, putative inhibitors should possess a high level of
selectivity for ALR2 over the related aldehyde reductase (ALR1). We investigated
the selectivity-determining features by gradually mapping the residues deviating
between the binding pockets of ALR1 and ALR2 into the ALR2 binding pocket. The
resulting mutational constructs of ALR2 (eight point mutations and one double
mutant) were probed for their influence towards ligand selectivity by X-ray
structure analysis of the corresponding complexes and isothermal titration
calorimetry (ITC). The binding properties of these mutants were evaluated using
a ligand set of zopolrestat, a related uracil derivative, IDD388, IDD393,
sorbinil, fidarestat and tolrestat. Our study revealed induced-fit adaptations
within the mutated binding site as an essential prerequisite for ligand
accommodation related to the selectivity discrimination of the ligands. However,
our study also highlights the limits of the present understanding of
protein-ligand interactions. Interestingly, binding site mutations not involved
in any direct interaction to the ligands in various cases show significant
effects towards their binding thermodynamics. Furthermore, our results suggest
the binding site residues deviating between ALR1 and ALR2 influence ligand
affinity in a complex interplay, presumably involving changes of dynamic
properties and differences of the solvation/desolvation balance upon ligand
binding.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Comparative stereo representations of the related
ALR1 (a) and ALR2 (b) inhibitor binding pockets.
|
 |
Figure 2.
Fig. 2. Chemical formulae of the ALR2 inhibitors investigated
in this study.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
379,
991-1016)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.J.Falconer,
A.Penkova,
I.Jelesarov,
and
B.M.Collins
(2010).
Survey of the year 2008: applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
23,
395-413.
|
 |
|
|
|
|
 |
O.A.Barski,
S.M.Tipparaju,
and
A.Bhatnagar
(2008).
The aldo-keto reductase superfamily and its role in drug metabolism and detoxification.
|
| |
Drug Metab Rev,
40,
553-624.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

