 |
PDBsum entry 2p3v

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Thermotoga maritima impase tm1415
|
|
Structure:
|
 |
Inositol-1-monophosphatase. Chain: a, b, c, d. Synonym: impase, inositol-1-phosphatase, i-1-pase. Engineered: yes
|
|
Source:
|
 |
Thermotoga maritima. Organism_taxid: 243274. Strain: msb8. Gene: suhb. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.178
|
R-free:
|
0.263
|
|
|
Authors:
|
 |
K.A.Stieglitz,M.F.Roberts,W.Li,B.Stec
|
|
Key ref:
|
 |
K.A.Stieglitz
et al.
(2007).
Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J,
274,
2461-2469.
PubMed id:
|
 |
|
Date:
|
 |
|
09-Mar-07
|
Release date:
|
24-Apr-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O33832
(BSUHB_THEMA) -
Fructose-1,6-bisphosphatase/inositol-1-monophosphatase from Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
256 a.a.
254 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.11
- fructose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
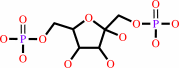 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
+
|
H2O
|
=
|
beta-D-fructose 6-phosphate
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.25
- inositol-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a myo-inositol phosphate + H2O = myo-inositol + phosphate
|
 |
 |
 |
 |
 |
myo-inositol phosphate
|
+
|
H2O
|
=
|
myo-inositol
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Febs J
274:2461-2469
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
|
|
K.A.Stieglitz,
M.F.Roberts,
W.Li,
B.Stec.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the first tetrameric inositol monophosphatase (IMPase) has been
solved. This enzyme, from the eubacterium Thermotoga maritima, similarly to its
archaeal homologs exhibits dual specificity with both IMPase and
fructose-1,6-bisphosphatase activities. The tetrameric structure of this
unregulated enzyme is similar, in its quaternary assembly, to the allosterically
regulated tetramer of fructose-1,6-bisphosphatase. The individual dimers are
similar to the human IMPase. Additionally, the structures of two crystal forms
of IMPase show significant differences. In the first crystal form, the
tetrameric structure is symmetrical, with the active site loop in each subunit
folded into a beta-hairpin conformation. The second form is asymmetrical and
shows an unusual structural change. Two of the subunits have the active site
loop folded into a beta-hairpin structure, whereas in the remaining two subunits
the same loop adopts an alpha-helical conformation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.Li,
K.A.Stieglitz,
A.L.Shrout,
Y.Wei,
R.M.Weis,
B.Stec,
and
M.F.Roberts
(2010).
Mobile loop mutations in an archaeal inositol monophosphatase: modulating three-metal ion assisted catalysis and lithium inhibition.
|
| |
Protein Sci,
19,
309-318.
|
 |
|
|
|
|
 |
P.V.Burra,
Y.Zhang,
A.Godzik,
and
B.Stec
(2009).
Global distribution of conformational states derived from redundant models in the PDB points to non-uniqueness of the protein structure.
|
| |
Proc Natl Acad Sci U S A,
106,
10505-10510.
|
 |
|
|
|
|
 |
R.H.Michell
(2008).
Inositol derivatives: evolution and functions.
|
| |
Nat Rev Mol Cell Biol,
9,
151-161.
|
 |
|
|
|
|
 |
A.K.Brown,
G.Meng,
H.Ghadbane,
D.J.Scott,
L.G.Dover,
J.Nigou,
G.S.Besra,
and
K.Fütterer
(2007).
Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+.
|
| |
BMC Struct Biol,
7,
55.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

