 |
PDBsum entry 2ohy

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase, transferase
|
PDB id
|
|

|

|
2ohy
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.3.1.23
- tyrosine ammonia-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tyrosine = (E)-4-coumarate + NH4+
|
 |
 |
 |
 |
 |
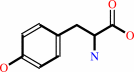 L-tyrosine
L-tyrosine
|
=
|
(E)-4-coumarate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
MIO
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.3.6
- tyrosine 2,3-aminomutase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tyrosine = 3-amino-3-(4-hydroxyphenyl)propanoate
|
 |
 |
 |
 |
 |
 L-tyrosine
L-tyrosine
|
=
|
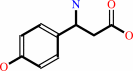 3-amino-3-(4-hydroxyphenyl)propanoate
3-amino-3-(4-hydroxyphenyl)propanoate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
MIO
|
 |
 |
 |
 |
 |
Cobalamin
|
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
46:7205-7214
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway.
|
|
C.V.Christianson,
T.J.Montavon,
S.G.Van Lanen,
B.Shen,
S.D.Bruner.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The SgcC4 l-tyrosine 2,3-aminomutase (SgTAM) catalyzes the formation of
(S)-beta-tyrosine in the biosynthetic pathway of the enediyne antitumor
antibiotic C-1027. SgTAM is homologous to the histidine ammonia lyase family of
enzymes whose activity is dependent on the methylideneimidazole-5-one (MIO)
cofactor. Unlike the lyase enzymes, SgTAM catalyzes additional chemical
transformations resulting in an overall stereospecific 1,2-amino shift in the
substrate l-tyrosine to generate (S)-beta-tyrosine. Previously, we provided
kinetic, spectroscopic, and mutagenesis data supporting the presence of MIO in
the active site of SgTAM [Christenson, S. D.; Wu, W.; Spies, A.; Shen, B.; and
Toney, M. D. (2003) Biochemistry 42, 12708-12718]. Here we report the first
X-ray crystal structure of an MIO-containing aminomutase, SgTAM, and confirm the
structural homology of SgTAM to ammonia lyases. Comparison of the structure of
SgTAM to the l-tyrosine ammonia lyase from Rhodobacter sphaeroides provides
insight into the structural basis for aminomutase activity. The results show
that SgTAM has a closed active site well suited to retain ammonia and minimize
the formation of lyase elimination products. The amino acid determinants for
substrate recognition and catalysis can be predicted from the structure, setting
the framework for detailed mechanistic investigations.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.J.Turner
(2011).
Ammonia lyases and aminomutases as biocatalysts for the synthesis of α-amino and β-amino acids.
|
| |
Curr Opin Chem Biol,
15,
234-240.
|
 |
|
|
|
|
 |
B.Wu,
W.Szymański,
H.J.Wijma,
C.G.Crismaru,
S.de Wildeman,
G.J.Poelarends,
B.L.Feringa,
and
D.B.Janssen
(2010).
Engineering of an enantioselective tyrosine aminomutase by mutation of a single active site residue in phenylalanine aminomutase.
|
| |
Chem Commun (Camb),
46,
8157-8159.
|
 |
|
|
|
|
 |
H.A.Cooke,
and
S.D.Bruner
(2010).
Probing the active site of MIO-dependent aminomutases, key catalysts in the biosynthesis of beta-amino acids incorporated in secondary metabolites.
|
| |
Biopolymers,
93,
802-810.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Du,
and
L.Lou
(2010).
PKS and NRPS release mechanisms.
|
| |
Nat Prod Rep,
27,
255-278.
|
 |
|
|
|
|
 |
Z.X.Liang
(2010).
Complexity and simplicity in the biosynthesis of enediyne natural products.
|
| |
Nat Prod Rep,
27,
499-528.
|
 |
|
|
|
|
 |
B.Wu,
W.Szymanski,
P.Wietzes,
S.de Wildeman,
G.J.Poelarends,
B.L.Feringa,
and
D.B.Janssen
(2009).
Enzymatic Synthesis of Enantiopure alpha- and beta-Amino Acids by Phenylalanine Aminomutase-Catalysed Amination of Cinnamic Acid Derivatives.
|
| |
Chembiochem,
10,
338-344.
|
 |
|
|
|
|
 |
D.Krug,
and
R.Müller
(2009).
Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF.
|
| |
Chembiochem,
10,
741-750.
|
 |
|
|
|
|
 |
H.A.Cooke,
C.V.Christianson,
and
S.D.Bruner
(2009).
Structure and chemistry of 4-methylideneimidazole-5-one containing enzymes.
|
| |
Curr Opin Chem Biol,
13,
460-468.
|
 |
|
|
|
|
 |
S.Lin,
S.G.Van Lanen,
and
B.Shen
(2009).
A free-standing condensation enzyme catalyzing ester bond formation in C-1027 biosynthesis.
|
| |
Proc Natl Acad Sci U S A,
106,
4183-4188.
|
 |
|
|
|
|
 |
L.Wang,
A.Gamez,
H.Archer,
E.E.Abola,
C.N.Sarkissian,
P.Fitzpatrick,
D.Wendt,
Y.Zhang,
M.Vellard,
J.Bliesath,
S.M.Bell,
J.F.Lemontt,
C.R.Scriver,
and
R.C.Stevens
(2008).
Structural and biochemical characterization of the therapeutic Anabaena variabilis phenylalanine ammonia lyase.
|
| |
J Mol Biol,
380,
623-635.
|
 |
|
|
|
|
 |
S.G.Van Lanen,
T.J.Oh,
W.Liu,
E.Wendt-Pienkowski,
and
B.Shen
(2007).
Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis.
|
| |
J Am Chem Soc,
129,
13082-13094.
|
 |
|
|
|
|
 |
S.Lin,
S.G.Van Lanen,
and
B.Shen
(2007).
Regiospecific chlorination of (S)-beta-tyrosyl-S-carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027.
|
| |
J Am Chem Soc,
129,
12432-12438.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

