 |
PDBsum entry 2odr

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 491 a.a.
491 a.a.
|
 |

|

|

|

|

|

|

|

 448 a.a.
448 a.a.
|
 |

|

|

|

|

|

|

|
 451 a.a.
451 a.a.
|
 |

|

|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Methanococcus maripaludis phosphoseryl-tRNA synthetase
|
|
Structure:
|
 |
Phosphoseryl-tRNA synthetase. Chain: a. Engineered: yes. Phosphoseryl-tRNA synthetase. Chain: b. Engineered: yes. Phosphoseryl-tRNA synthetase. Chain: c. Engineered: yes.
|
|
Source:
|
 |
Methanococcus maripaludis. Organism_taxid: 267377. Strain: s2. Gene: mmp0688. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
3.23Å
|
R-factor:
|
0.292
|
R-free:
|
0.306
|
|
|
Authors:
|
 |
T.A.Steitz,S.Kamtekar
|
Key ref:

|
 |
S.Kamtekar
et al.
(2007).
Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc Natl Acad Sci U S A,
104,
2620-2625.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
26-Dec-06
|
Release date:
|
13-Feb-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q6LZE1
(SEPS_METMP) -
O-phosphoserine--tRNA(Cys) ligase from Methanococcus maripaludis (strain S2 / LL)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
537 a.a.
491 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.6.1.1.27
- O-phosphoserine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Cys) + O-phospho-L-serine + ATP = O-phospho-L-seryl-tRNA(Cys) + AMP + diphosphate
|
 |
 |
 |
 |
 |
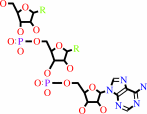 tRNA(Cys)
tRNA(Cys)
|
+
|
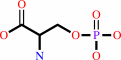 O-phospho-L-serine
O-phospho-L-serine
|
+
|
 ATP
ATP
|
=
|
O-phospho-L-seryl-tRNA(Cys)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
104:2620-2625
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
|
|
S.Kamtekar,
M.J.Hohn,
H.S.Park,
M.Schnitzbauer,
A.Sauerwald,
D.Söll,
T.A.Steitz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A number of archaeal organisms generate Cys-tRNA(Cys) in a two-step pathway,
first charging phosphoserine (Sep) onto tRNA(Cys) and subsequently converting it
to Cys-tRNA(Cys). We have determined, at 3.2-A resolution, the structure of the
Methanococcus maripaludis phosphoseryl-tRNA synthetase (SepRS), which catalyzes
the first step of this pathway. The structure shows that SepRS is a class II,
alpha(4) synthetase whose quaternary structure arrangement of subunits closely
resembles that of the heterotetrameric (alphabeta)(2) phenylalanyl-tRNA
synthetase (PheRS). Homology modeling of a tRNA complex indicates that, in
contrast to PheRS, a single monomer in the SepRS tetramer may recognize both the
acceptor terminus and anticodon of a tRNA substrate. Using a complex with
tungstate as a marker for the position of the phosphate moiety of Sep, we
suggest that SepRS and PheRS bind their respective amino acid substrates in
dissimilar orientations by using different residues.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. The active site of M. maripaludis SepRS compared
with that of T. thermophilus PheRS (PDB ID code 1JJC) (29). (A)
The catalytic domain of one monomer of SepRS (cyan) is shown
superposed on a catalytic domain of PheRS (gray). This
superposition is used in all images. Residues mutated in SepRS
are shown in stick representation. Residue labels in all images
are colored to indicate the severity of the mutant phenotype,
and corresponding residue numbers in PheRS are shown in
brackets. (B) Three T. thermophilus PheRS residues that interact
with the AMP moiety are shown in gray. The corresponding
residues in M. maripaludis SepRS are conserved (green). (C) Four
T. thermophilus PheRS residues that form a binding pocket for
the phenylalanyl side chain are shown in gray. The corresponding
residues in M. maripaludis SepRS (green) are incompatible with
the binding of phenylalanine. (D) Residues in M. maripaludis
SepRS that may interact with the phosphate moiety of
phosphoserine. Difference electron density, caused by incubating
a crystal with sodium tungstate and contoured at 6  , is
shown as a mesh and superimposed on the structure both here and
in F. (E and F) Side-by-side comparison of the interactions of
T. thermophilus PheRS with phenylalanyl-adenylate and M.
maripaludis SepRS with modeled phosphoseryl-adenylate. The
side-chain conformations of M. maripaludis SepRS residues H186,
T188, and R216 have been altered from the apo structure to
accommodate the phosphoseryl moiety. The phosphoseryl phosphate
group occupies a hydrophilic pocket, and its position overlaps
the tungstate difference electron density. , is
shown as a mesh and superimposed on the structure both here and
in F. (E and F) Side-by-side comparison of the interactions of
T. thermophilus PheRS with phenylalanyl-adenylate and M.
maripaludis SepRS with modeled phosphoseryl-adenylate. The
side-chain conformations of M. maripaludis SepRS residues H186,
T188, and R216 have been altered from the apo structure to
accommodate the phosphoseryl moiety. The phosphoseryl phosphate
group occupies a hydrophilic pocket, and its position overlaps
the tungstate difference electron density.
|
 |
Figure 3.
Fig. 3. tRNA^Phe homology modeled onto the structure of
SepRS by using the superposition of the catalytic domains of M.
maripaludis SepRS and T. thermophilus PheRS (19) illustrated in
Fig. 1B. The tRNAs are shown as white/gray surfaces; each
polypeptide chain of SepRS is colored differently, and
interactions between SepRS and one of the tRNAs are labeled.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Finarov,
N.Moor,
N.Kessler,
L.Klipcan,
and
M.G.Safro
(2010).
Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns.
|
| |
Structure,
18,
343-353.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Banerjee,
S.Chen,
K.Dare,
M.Gilreath,
M.Praetorius-Ibba,
M.Raina,
N.M.Reynolds,
T.Rogers,
H.Roy,
S.S.Yadavalli,
and
M.Ibba
(2010).
tRNAs: cellular barcodes for amino acids.
|
| |
FEBS Lett,
584,
387-395.
|
 |
|
|
|
|
 |
C.M.Zhang,
C.Liu,
S.Slater,
and
Y.M.Hou
(2008).
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys).
|
| |
Nat Struct Mol Biol,
15,
507-514.
|
 |
|
|
|
|
 |
J.Yuan,
K.Sheppard,
and
D.Söll
(2008).
Amino acid modifications on tRNA.
|
| |
Acta Biochim Biophys Sin (Shanghai),
40,
539-553.
|
 |
|
|
|
|
 |
K.Sheppard,
J.Yuan,
M.J.Hohn,
B.Jester,
K.M.Devine,
and
D.Söll
(2008).
From one amino acid to another: tRNA-dependent amino acid biosynthesis.
|
| |
Nucleic Acids Res,
36,
1813-1825.
|
 |
|
|
|
|
 |
L.Klipcan,
I.Levin,
N.Kessler,
N.Moor,
I.Finarov,
and
M.Safro
(2008).
The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase.
|
| |
Structure,
16,
1095-1104.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.I.Hauenstein,
Y.M.Hou,
and
J.J.Perona
(2008).
The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity.
|
| |
J Biol Chem,
283,
21997-22006.
|
 |
|
|
|
|
 |
J.M.Kavran,
S.Gundllapalli,
P.O'Donoghue,
M.Englert,
D.Söll,
and
T.A.Steitz
(2007).
Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
|
| |
Proc Natl Acad Sci U S A,
104,
11268-11273.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
| |

