 |
PDBsum entry 2o7c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of l-methionine-lyase from pseudomonas
|
|
Structure:
|
 |
Methionine gamma-lyase. Chain: a, b, c, d. Synonym: l-methioninase. Engineered: yes
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.204
|
R-free:
|
0.224
|
|
|
Authors:
|
 |
S.Misaki,A.Takimoto,T.Takakura,T.Yoshioka,M.Yamashita,T.Tamura, H.Tanaka,K.Inagaki
|
|
Key ref:
|
 |
D.Kudou
et al.
(2007).
Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution.
J Biochem (tokyo),
141,
535-544.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Dec-06
|
Release date:
|
11-Dec-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P13254
(MEGL_PSEPU) -
L-methionine gamma-lyase from Pseudomonas putida
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
398 a.a.
398 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.4.1.11
- methionine gamma-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionine + H2O = methanethiol + 2-oxobutanoate + NH4+
|
 |
 |
 |
 |
 |
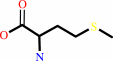 L-methionine
L-methionine
|
+
|
H2O
|
=
|
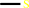 methanethiol
methanethiol
|
+
|
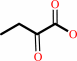 2-oxobutanoate
2-oxobutanoate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.2
- homocysteine desulfhydrase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homocysteine + H2O = 2-oxobutanoate + hydrogen sulfide + NH4+ + H+
|
 |
 |
 |
 |
 |
 L-homocysteine
L-homocysteine
|
+
|
H2O
|
=
|
 2-oxobutanoate
2-oxobutanoate
|
+
|
hydrogen sulfide
|
+
|
NH4(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biochem (tokyo)
141:535-544
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution.
|
|
D.Kudou,
S.Misaki,
M.Yamashita,
T.Tamura,
T.Takakura,
T.Yoshioka,
S.Yagi,
R.M.Hoffman,
A.Takimoto,
N.Esaki,
K.Inagaki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
l-Methionine gamma-lyase (EC 4.4.1.11, MGL_Pp) from Pseudomonas putida is a
multifunctional enzyme, which belongs to the gamma-family of
pyridoxal-5'-phosphate (PLP) dependent enzymes. In this report, we demonstrate
that the three-dimensional structure of MGL_Pp has been completely solved by the
molecular replacement method to an R-factor of 20.4% at 1.8 A resolution.
Detailed information of the overall structure of MGL_Pp supplies a clear picture
of the substrate- and PLP-binding pockets. Tyr59 and Arg61 of neighbouring
subunits, which are strongly conserved in other gamma-family enzymes, contact
the phosphate group of PLP. These residues are important as the main anchor
within the active site. Lys240, Asp241 and Arg61 of one partner monomer and
Tyr114 and Cys116 of the other partner monomer form a hydrogen-bond network in
the MGL active site which is specific for MGLs. It is also suggested that
electrostatic interactions at the subunit interface are involved in the
stabilization of the structural conformation. The detailed structure will
facilitate the development of MGL_Pp as an anticancer drug.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.S.El-Sayed
(2011).
Purification and characterization of a new L-methioninase from solid cultures of Aspergillus flavipes.
|
| |
J Microbiol,
49,
130-140.
|
 |
|
|
|
|
 |
A.S.El-Sayed
(2010).
Microbial L-methioninase: production, molecular characterization, and therapeutic applications.
|
| |
Appl Microbiol Biotechnol,
86,
445-467.
|
 |
|
|
|
|
 |
A.Nikulin,
S.Revtovich,
E.Morozova,
N.Nevskaya,
S.Nikonov,
M.Garber,
and
T.Demidkina
(2008).
High-resolution structure of methionine gamma-lyase from Citrobacter freundii.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
211-218.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Kudou,
S.Misaki,
M.Yamashita,
T.Tamura,
N.Esaki,
and
K.Inagaki
(2008).
The role of cysteine 116 in the active site of the antitumor enzyme L-methionine gamma-lyase from Pseudomonas putida.
|
| |
Biosci Biotechnol Biochem,
72,
1722-1730.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

