 |
PDBsum entry 2nsf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.2.1.4
- maleylpyruvate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-maleylpyruvate = 3-fumarylpyruvate
|
 |
 |
 |
 |
 |
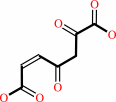 3-maleylpyruvate
3-maleylpyruvate
|
=
|
 3-fumarylpyruvate
3-fumarylpyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:16288-15294
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal Structures and Site-directed Mutagenesis of a Mycothiol-dependent Enzyme Reveal a Novel Folding and Molecular Basis for Mycothiol-mediated Maleylpyruvate Isomerization.
|
|
R.Wang,
Y.J.Yin,
F.Wang,
M.Li,
J.Feng,
H.M.Zhang,
J.P.Zhang,
S.J.Liu,
W.R.Chang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mycothiol (MSH) is the major low molecular mass thiols in many Gram-positive
bacteria such as Mycobacterium tuberculosis and Corynebacterium glutamicum. The
physiological roles of MSH are believed to be equivalent to those of GSH in
Gram-negative bacteria, but current knowledge of MSH is limited to
detoxification of alkalating chemicals and protection from host cell
defense/killing systems. Recently, an MSH-dependent maleylpyruvate isomerase
(MDMPI) was discovered from C. glutamicum, and this isomerase represents one
example of many putative MSH-dependent enzymes that take MSH as cofactor. In
this report, fourteen mutants of MDMPI were generated. The wild type and mutant
(H52A) MDMPIs were crystallized and their structures were solved at 1.75 and
2.05A resolution, respectively. The crystal structures reveal that this enzyme
contains a divalent metal-binding domain and a C-terminal domain possessing a
novel folding pattern (alphabetaalphabetabetaalpha fold). The divalent
metal-binding site is composed of residues His(52), Glu(144), and His(148) and
is located at the bottom of a surface pocket. Combining the structural and
site-directed mutagenesis studies, it is proposed that this surface pocket
including the metal ion and MSH moiety formed the putative catalytic center.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIGURE 4. The side chain of Arg^222 (NH1) at the C-terminal
domain interacts with Asp^151 (OD2) of the N-terminal domain
through a salt bridge. The N-terminal domain is shown in yellow,
C-terminal domain in cyan, and interdomain coil in gray. Asp^151
and Arg^222 are represented in stick model (carbon atoms are
colored the same as the domain they locate at) and the metal ion
by a sphere (gray). Salt bridge is shown by dashed line. The
figure was created by PyMol.
|
 |
Figure 6.
FIGURE 6. The stereo view of the conformation differences
in the putative active pocket between the wild type and the
mutant (H52A) MDMPI. Mutant H52A is shown in purple and native
MDMPI in green. Conformation differences were found at the side
chains of Arg^82 and Trp^44. In H52A the metal ion was missing
and a glycerol occupied the position of the metal ion, and the
location of SO^2–[4] also changed. Furthermore, there is no
obvious conformation changes observed, when compared with that
of the wild type. The image was created by PyMol.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
16288-15294)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.T.Liu,
Y.Xu,
H.Liu,
S.Luo,
Y.J.Yin,
S.J.Liu,
and
N.Y.Zhou
(2011).
Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038.
|
| |
Appl Microbiol Biotechnol,
90,
671-678.
|
 |
|
|
|
|
 |
D.R.Cooper,
K.Grelewska,
C.Y.Kim,
A.Joachimiak,
and
Z.S.Derewenda
(2010).
The structure of DinB from Geobacillus stearothermophilus: a representative of a unique four-helix-bundle superfamily.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
219-224.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.J.Yin,
B.J.Wang,
C.Y.Jiang,
Y.M.Luo,
J.H.Jin,
and
S.J.Liu
(2010).
Identification and quantification of mycothiol in Actinobacteria by a novel enzymatic method.
|
| |
Appl Microbiol Biotechnol,
88,
1393-1401.
|
 |
|
|
|
|
 |
E.Ordóñez,
K.Van Belle,
G.Roos,
S.De Galan,
M.Letek,
J.A.Gil,
L.Wyns,
L.M.Mateos,
and
J.Messens
(2009).
Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange.
|
| |
J Biol Chem,
284,
15107-15116.
|
 |
|
|
|
|
 |
G.L.Newton,
N.Buchmeier,
and
R.C.Fahey
(2008).
Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria.
|
| |
Microbiol Mol Biol Rev,
72,
471-494.
|
 |
|
|
|
|
 |
K.Nagata,
J.Ohtsuka,
M.Takahashi,
A.Asano,
H.Iino,
A.Ebihara,
and
M.Tanokura
(2008).
Crystal structure of TTHA0303 (TT2238), a four-helix bundle protein with an exposed histidine triad from Thermus thermophilus HB8 at 2.0 A.
|
| |
Proteins,
70,
1103-1107.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.K.Jothivasan,
and
C.J.Hamilton
(2008).
Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes.
|
| |
Nat Prod Rep,
25,
1091-1117.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

