 |
PDBsum entry 2j2c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of human cytosolic 5'-nucleotidase ii (nt5c2, cn-ii)
|
|
Structure:
|
 |
Cytosolic purine 5'-nucleotidase. Chain: a. Fragment: residues 1-536. Synonym: 5'-nucleotidase cytosolic ii, cytosolic purine 5'- nucleotidase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_cell_line: rosetta2(de).
|
|
Biol. unit:
|
 |
Monomer (from PDB file)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.154
|
R-free:
|
0.184
|
|
|
Authors:
|
 |
K.Wallden,P.Stenmark,C.Arrowsmith,H.Berglund,R.Busam,R.Collins, A.Edwards,M.Ehn,S.Flodin,A.Flores,S.Graslund,M.Hammarstrom, B.M.Hallberg,L.Holmberg Schiavone,M.Hogbom,T.Karlberg,T.Kotenyova, P.Loppnau,A.Magnusdottir,P.Nilsson-Ehle,T.Nyman,D.Ogg,C.Persson, J.Sagemark,M.Sundstrom,J.Uppenberg,A.G.Thorsell,S.Van Den Berg, J.Weigelt,M.Welin,P.Nordlund
|
Key ref:

|
 |
K.Walldén
et al.
(2007).
Crystal structure of human cytosolic 5'-nucleotidase II: insights into allosteric regulation and substrate recognition.
J Biol Chem,
282,
17828-17836.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Aug-06
|
Release date:
|
19-Sep-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49902
(5NTC_HUMAN) -
Cytosolic purine 5'-nucleotidase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
561 a.a.
470 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.1.77
- nucleoside phosphotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside + a ribonucleoside 5'-phosphate = a ribonucleoside + a 2'-deoxyribonucleoside 5'-phosphate
|
 |
 |
 |
 |
 |
2'-deoxyribonucleoside
|
+
|
ribonucleoside 5'-phosphate
|
=
|
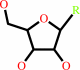 ribonucleoside
ribonucleoside
|
+
|
2'-deoxyribonucleoside 5'-phosphate
Bound ligand (Het Group name = )
matches with 60.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.5
- 5'-nucleotidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
|
 |
 |
 |
 |
 |
ribonucleoside 5'-phosphate
|
+
|
H2O
|
=
|
ribonucleoside
Bound ligand (Het Group name = )
matches with 60.00% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.3.99
- IMP-specific 5'-nucleotidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + H2O = inosine + phosphate
|
 |
 |
 |
 |
 |
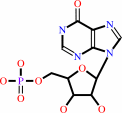 IMP
IMP
|
+
|
H2O
|
=
|
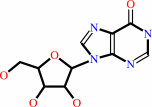 inosine
inosine
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent metal cation
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:17828-17836
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human cytosolic 5'-nucleotidase II: insights into allosteric regulation and substrate recognition.
|
|
K.Walldén,
P.Stenmark,
T.Nyman,
S.Flodin,
S.Gräslund,
P.Loppnau,
V.Bianchi,
P.Nordlund.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cytosolic 5'-nucleotidase II catalyzes the dephosphorylation of 6-hydroxypurine
nucleoside 5'-monophosphates and regulates the IMP and GMP pools within the
cell. It possesses phosphotransferase activity and thereby also catalyzes the
reverse reaction. Both reactions are allosterically activated by adenine-based
nucleotides and 2,3-bisphosphoglycerate. We have solved structures of cytosolic
5'-nucleotidase II as native protein (2.2 Angstrom) and in complex with
adenosine (1.5 Angstrom) and beryllium trifluoride (2.15 Angstrom) The
tetrameric enzyme is structurally similar to enzymes of the haloacid
dehalogenase (HAD) superfamily, including mitochondrial
5'(3')-deoxyribonucleotidase and cytosolic 5'-nucleotidase III but possesses
additional regulatory regions that contain two allosteric effector sites. At
effector site 1 located near a subunit interface we modeled diadenosine
tetraphosphate with one adenosine moiety in each subunit. This efficiently glues
the tetramer subunits together in pairs. The model shows why diadenosine
tetraphosphate but not diadenosine triphosphate activates the enzyme and
supports a role for cN-II during apoptosis when the level of diadenosine
tetraphosphate increases. We have also modeled 2,3-bisphosphoglycerate in
effector site 1 using one phosphate site from each subunit. By comparing the
structure of cytosolic 5'-nucleotidase II with that of mitochondrial
5'(3')-deoxyribonucleotidase in complex with dGMP, we identified residues
involved in substrate recognition.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
FIGURE 5. Effector sites 1 and 2 of cN-II with bound
adenosine and sulfate. Omit F[o] - F[c] maps are covering
adenosine with  level of 4.5 in
effector site 1 and level of 4.5 in
effector site 1 and  level of 3 in effector
site 2. A, stereo image of adenosine bound in effector site 1,
with adenosine and amino acid residues colored in turquoise. B,
stereo image of effector site 1 from two adjacent subunits
connected through interface A. Salt bridges are indicated by the
dotted lines. Nitrogens are shown in blue, oxygens in red, and
sulfur in yellow. C, stereo image of adenosine bound in effector
site 2. Note that the ribose moiety is disordered so that the
electron density cannot confirm its location. Waters are colored
in red. Polar atoms are color-coded as following: nitrogen in
blue, oxygens in red, and sulfur in yellow. level of 3 in effector
site 2. A, stereo image of adenosine bound in effector site 1,
with adenosine and amino acid residues colored in turquoise. B,
stereo image of effector site 1 from two adjacent subunits
connected through interface A. Salt bridges are indicated by the
dotted lines. Nitrogens are shown in blue, oxygens in red, and
sulfur in yellow. C, stereo image of adenosine bound in effector
site 2. Note that the ribose moiety is disordered so that the
electron density cannot confirm its location. Waters are colored
in red. Polar atoms are color-coded as following: nitrogen in
blue, oxygens in red, and sulfur in yellow.
|
 |
Figure 7.
FIGURE 7. Two subunits of the tetrameric cN-II with three
positively charged regions (K(25)KYRR), (K(359)SKKRQ), and
(Q(420)RRIKK) shown in orange. Adenosines (white), sulfates
(yellow and red), and magnesium (yellow) are also shown.
Nitrogens are shown in blue and oxygens in red. The C-terminal
end of the structure (residue 488) is marked C.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
17828-17836)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.A.Meyer,
J.Wang,
L.E.Hogan,
J.J.Yang,
S.Dandekar,
J.P.Patel,
Z.Tang,
P.Zumbo,
S.Li,
J.Zavadil,
R.L.Levine,
T.Cardozo,
S.P.Hunger,
E.A.Raetz,
W.E.Evans,
D.J.Morrison,
C.E.Mason,
and
W.L.Carroll
(2013).
Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia.
|
| |
Nat Genet,
45,
290-294.
|
 |
|
|
|
|
 |
K.Walldén,
and
P.Nordlund
(2011).
Structural Basis for the Allosteric Regulation and Substrate Recognition of Human Cytosolic 5'-Nucleotidase II.
|
| |
J Mol Biol,
408,
684-696.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Pesi,
S.Allegrini,
M.G.Careddu,
D.N.Filoni,
M.Camici,
and
M.G.Tozzi
(2010).
Active and regulatory sites of cytosolic 5'-nucleotidase.
|
| |
FEBS J,
277,
4863-4872.
|
 |
|
|
|
|
 |
P.Mirandola,
G.Gobbi,
I.Sponzilli,
C.Malinverno,
A.Cavazzoni,
R.Alfieri,
P.G.Petronini,
and
M.Vitale
(2009).
TRAIL-induced apoptosis of FHIT-negative lung cancer cells is inhibited by FHIT re-expression.
|
| |
J Cell Physiol,
220,
492-498.
|
 |
|
|
|
|
 |
J.Weigelt,
L.D.McBroom-Cerajewski,
M.Schapira,
Y.Zhao,
C.H.Arrowsmith,
and
C.H.Arrowmsmith
(2008).
Structural genomics and drug discovery: all in the family.
|
| |
Curr Opin Chem Biol,
12,
32-39.
|
 |
|
|
|
|
 |
L.Li,
B.Fridley,
K.Kalari,
G.Jenkins,
A.Batzler,
S.Safgren,
M.Hildebrandt,
M.Ames,
D.Schaid,
and
L.Wang
(2008).
Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression.
|
| |
Cancer Res,
68,
7050-7058.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

