
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 2 more)
459 a.a.
(+ 2 more)
459 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 2 more)
258 a.a.
(+ 2 more)
258 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of methanol:cobalamin methyltransferase complex mtabc from methanosarcina barkeri
|
|
Structure:
|
 |
Methyltransferase 1. Chain: a, c, e, g, i, k, m, o. Synonym: mtab. Methyltransferase 1. Chain: b, d, f, h, j, l, n, p. Synonym: mtac. Ec: 2.1.1.90
|
|
Source:
|
 |
Methanosarcina barkeri. Organism_taxid: 2208. Strain: fuaro (dsm 804). Strain: fuaro (dsm 804)
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.184
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
C.H.Hagemeier,M.Kruer,R.K.Thauer,E.Warkentin,U.Ermler
|
Key ref:

|
 |
C.H.Hagemeier
et al.
(2006).
Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex.
Proc Natl Acad Sci U S A,
103,
18917-18922.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Aug-06
|
Release date:
|
21-Nov-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.2.1.1.90
- methanol--corrinoid protein Co-methyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Co(I)-[methanol-specific corrinoid protein] + methanol + H+ = methyl- Co(III)-[methanol-specific corrinoid protein] + H2O
|
 |
 |
 |
 |
 |
Co(I)-[methanol-specific corrinoid protein]
|
+
|
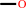 methanol
methanol
|
+
|
H(+)
|
=
|
methyl- Co(III)-[methanol-specific corrinoid protein]
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:18917-18922
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex.
|
|
C.H.Hagemeier,
M.Krer,
R.K.Thauer,
E.Warkentin,
U.Ermler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Some methanogenic and acetogenic microorganisms have the catalytic capability to
cleave heterolytically the C O bond of methanol. To obtain insight into the
elusive enzymatic mechanism of this challenging chemical reaction we have
investigated the methanol-activating MtaBC complex from Methanosarcina barkeri
composed of the zinc-containing MtaB and the
5-hydroxybenzimidazolylcobamide-carrying MtaC subunits. Here we report the 2.5-A
crystal structure of this complex organized as a (MtaBC)(2) heterotetramer. MtaB
folds as a TIM barrel and contains a novel zinc-binding motif. Zinc(II) lies at
the bottom of a funnel formed at the C-terminal beta-barrel end and ligates to
two cysteinyl sulfurs (Cys-220 and Cys-269) and one carboxylate oxygen
(Glu-164). MtaC is structurally related to the cobalamin-binding domain of
methionine synthase. Its corrinoid cofactor at the top of the Rossmann domain
reaches deeply into the funnel of MtaB, defining a region between zinc(II) and
the corrinoid cobalt that must be the binding site for methanol. The active site
geometry supports a S(N)2 reaction mechanism, in which the C O bond in methanol
is activated by the strong electrophile zinc(II) and cleaved because of an
attack of the supernucleophile cob(I)amide. The environment of zinc(II) is
characterized by an acidic cluster that increases the charge density on the
zinc(II), polarizes methanol, and disfavors deprotonation of the methanol
hydroxyl group. Implications of the MtaBC structure for the second step of the
reaction, in which the methyl group is transferred to coenzyme M, are discussed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
The MtaBC structure. (A) In the (MtaBC)[2] heterotetramer (in
stereo) each MtaBC unit (MtaB in blue and MtaC in red) is
related to its partner unit (in light blue and red) by a twofold
noncrystallographic axis. The (MtaBC)[2] complex has a size of
62 Å × 55 Å × 52 Å and forms two
active sites separated by a distance of 38 Å. The
corrinoids are shown as stick models, and the zinc(II) ions are
depicted by green spheres. The contact loops between the two
active sites are highlighted in black. For oligomeric assembly
the interactions between the N-terminal extension of MtaC (in
green) and the counter MtaB are essential. (B) In the MtaBC unit
the Rossmann domain (orange) of MtaC is only loosely associated
with its helical domain (red) and MtaB (blue). MtaB is
subdivided into a TIM barrel core (dark blue) and a helical
layer (light blue). The active site is located between the
corrinoid cobalt and zinc(II).
|
 |
Figure 3.
Proposed mechanism of methanol activation. (A) Scheme of a
unique acidic cluster that flanks the zinc and methanol-binding
site. The acidic residues (in red) might play a crucial role in
polarizing zinc(II) and methanol. The unclear peak X was
tentatively assigned as a potassium ion. Methanol is modeled
into the protein, and its distance to the corrinoid Co is
estimated based on the distance between the zinc(II) and
methanol oxygen of 2 Å. (B) S[N]2 mechanism for the
methylation of 5-hydroxybenzimidazolyl cob(I)amide with
methanol. The methanol is activated by the strong electrophile
zinc(II) and attacked by the supernucleophile cob(I)amide.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Studenik,
S.Kreher,
and
G.Diekert
(2011).
The ether-cleaving methyltransferase of the strict anaerobe Acetobacterium dehalogenans: analysis of the zinc-binding site.
|
| |
FEMS Microbiol Lett,
318,
131-136.
|
 |
|
|
|
|
 |
M.A.Hossain,
M.A.Saeed,
G.Gryn'ova,
D.R.Powell,
and
J.Leszczynski
(2010).
Unusual complexes of trapped methanol with azacryptands.
|
| |
CrystEngComm,
2010,
4042-4044.
|
 |
|
|
|
|
 |
M.Rother,
and
J.A.Krzycki
(2010).
Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea.
|
| |
Archaea,
2010,
0.
|
 |
|
|
|
|
 |
A.Schilhabel,
S.Studenik,
M.Vödisch,
S.Kreher,
B.Schlott,
A.J.Pierik,
A.Y.Pierik,
and
G.Diekert
(2009).
The ether-cleaving methyltransferase system of the strict anaerobe Acetobacterium dehalogenans: analysis and expression of the encoding genes.
|
| |
J Bacteriol,
191,
588-599.
|
 |
|
|
|
|
 |
R.G.Matthews
(2009).
A love affair with vitamins.
|
| |
J Biol Chem,
284,
26217-26228.
|
 |
|
|
|
|
 |
T.Ferguson,
J.A.Soares,
T.Lienard,
G.Gottschalk,
and
J.A.Krzycki
(2009).
RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic archaea.
|
| |
J Biol Chem,
284,
2285-2295.
|
 |
|
|
|
|
 |
M.Koutmos,
R.Pejchal,
T.M.Bomer,
R.G.Matthews,
J.L.Smith,
and
M.L.Ludwig
(2008).
Metal active site elasticity linked to activation of homocysteine in methionine synthases.
|
| |
Proc Natl Acad Sci U S A,
105,
3286-3291.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.G.Matthews,
M.Koutmos,
and
S.Datta
(2008).
Cobalamin-dependent and cobamide-dependent methyltransferases.
|
| |
Curr Opin Struct Biol,
18,
658-666.
|
 |
|
|
|
|
 |
S.Datta,
M.Koutmos,
K.A.Pattridge,
M.L.Ludwig,
and
R.G.Matthews
(2008).
A disulfide-stabilized conformer of methionine synthase reveals an unexpected role for the histidine ligand of the cobalamin cofactor.
|
| |
Proc Natl Acad Sci U S A,
105,
4115-4120.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.W.Ragsdale,
and
E.Pierce
(2008).
Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation.
|
| |
Biochim Biophys Acta,
1784,
1873-1898.
|
 |
|
|
|
|
 |
S.W.Ragsdale
(2008).
Catalysis of methyl group transfers involving tetrahydrofolate and B(12).
|
| |
Vitam Horm,
79,
293-324.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

