 |
PDBsum entry 2hg2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2hg2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.2.1.21
- glycolaldehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glycolaldehyde + NAD+ + H2O = glycolate + NADH + 2 H+
|
 |
 |
 |
 |
 |
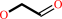 glycolaldehyde
glycolaldehyde
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
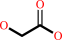 glycolate
glycolate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.2.1.22
- lactaldehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-lactaldehyde + NAD+ + H2O = (S)-lactate + NADH + 2 H+
|
 |
 |
 |
 |
 |
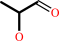 (S)-lactaldehyde
(S)-lactaldehyde
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
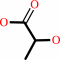 (S)-lactate
(S)-lactate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
366:481-493
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal Structure of Lactaldehyde Dehydrogenase from Escherichia coli and Inferences Regarding Substrate and Cofactor Specificity.
|
|
L.Di Costanzo,
G.A.Gomez,
D.W.Christianson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aldehyde dehydrogenases catalyze the oxidation of aldehyde substrates to the
corresponding carboxylic acids. Lactaldehyde dehydrogenase from Escherichia coli
(aldA gene product, P25553) is an NAD(+)-dependent enzyme implicated in the
metabolism of l-fucose and l-rhamnose. During the heterologous expression and
purification of taxadiene synthase from the Pacific yew, lactaldehyde
dehydrogenase from E. coli was identified as a minor (</=5%) side-product
subsequent to its unexpected crystallization. Accordingly, we now report the
serendipitous crystal structure determination of unliganded lactaldehyde
dehydrogenase from E. coli determined by the technique of multiple isomorphous
replacement using anomalous scattering at 2.2 A resolution. Additionally, we
report the crystal structure of the ternary enzyme complex with products lactate
and NADH at 2.1 A resolution, and the crystal structure of the enzyme complex
with NADPH at 2.7 A resolution. The structure of the ternary complex reveals
that the nicotinamide ring of the cofactor is disordered between two
conformations: one with the ring positioned in the active site in the so-called
hydrolysis conformation, and another with the ring extended out of the active
site into the solvent region, designated the out conformation. This represents
the first crystal structure of an aldehyde dehydrogenase-product complex. The
active site pocket in which lactate binds is more constricted than that of
medium-chain dehydrogenases such as the YdcW gene product of E. coli. The
structure of the binary complex with NADPH reveals the first view of the
structural basis of specificity for NADH: the negatively charged carboxylate
group of E179 destabilizes the binding of the 2'-phosphate group of NADPH
sterically and electrostatically, thereby accounting for the lack of enzyme
activity with this cofactor.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Pathways of l-fucose and l-rhamnose metabolism in E.
coli. Lactaldehyde dehydrogenase generates l-lactate, which is
converted to pyruvate for entry into the central metabolic
processes of the cell. Figure 1. Pathways of l-fucose and
l-rhamnose metabolism in E. coli. Lactaldehyde dehydrogenase
generates l-lactate, which is converted to pyruvate for entry
into the central metabolic processes of the cell.
|
 |
Figure 2.
Figure 2. The NAD^+-dependent oxidation of lactaldehyde to
lactate catalyzed by lactaldehyde dehydrogenase. Figure 2.
The NAD^+-dependent oxidation of lactaldehyde to lactate
catalyzed by lactaldehyde dehydrogenase.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Elsevier:
J Mol Biol
(2007,
366,
481-493)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Lee,
I.Kim,
J.Lee,
K.L.Lee,
B.Min,
and
C.Park
(2010).
Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12.
|
| |
J Bacteriol,
192,
4205-4214.
|
 |
|
|
|
|
 |
S.O.Kotchoni,
J.C.Jimenez-Lopez,
D.Gao,
V.Edwards,
E.W.Gachomo,
V.M.Margam,
and
M.J.Seufferheld
(2010).
Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features.
|
| |
PLoS One,
5,
e11516.
|
 |
|
|
|
|
 |
J.Crawford,
O.Grujic,
E.Bruic,
M.Czjzek,
M.E.Grigg,
and
M.J.Boulanger
(2009).
Structural characterization of the bradyzoite surface antigen (BSR4) from toxoplasma gondii, a unique addition to the surface antigen glycoprotein 1-related superfamily.
|
| |
J Biol Chem,
284,
9192-9198.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.G.Kim,
S.Lee,
O.S.Kwon,
S.Y.Park,
S.J.Lee,
B.J.Park,
and
K.J.Kim
(2009).
Redox-switch modulation of human SSADH by dynamic catalytic loop.
|
| |
EMBO J,
28,
959-968.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.N.Parkinson,
D.Vines,
P.C.Driscoll,
and
S.Djordjevic
(2008).
Crystal structures of PI3K-C2alpha PX domain indicate conformational change associated with ligand binding.
|
| |
BMC Struct Biol,
8,
13.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.S.Rodríguez-Zavala
(2008).
Enhancement of coenzyme binding by a single point mutation at the coenzyme binding domain of E. coli lactaldehyde dehydrogenase.
|
| |
Protein Sci,
17,
563-570.
|
 |
|
|
|
|
 |
S.Watanabe,
S.Piyanart,
and
K.Makino
(2008).
Metabolic fate of L-lactaldehyde derived from an alternative L-rhamnose pathway.
|
| |
FEBS J,
275,
5139-5149.
|
 |
|
|
|
|
 |
K.R.Hristova,
R.Schmidt,
A.Y.Chakicherla,
T.C.Legler,
J.Wu,
P.S.Chain,
K.M.Scow,
and
S.R.Kane
(2007).
Comparative transcriptome analysis of Methylibium petroleiphilum PM1 exposed to the fuel oxygenates methyl tert-butyl ether and ethanol.
|
| |
Appl Environ Microbiol,
73,
7347-7357.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

