 |
PDBsum entry 2hdh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2hdh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.35
- 3-hydroxyacyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3S)-3-hydroxyacyl-CoA + NAD+ = a 3-oxoacyl-CoA + NADH + H+
|
 |
 |
 |
 |
 |
 (3S)-3-hydroxyacyl-CoA
(3S)-3-hydroxyacyl-CoA
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
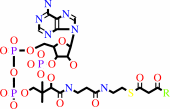 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:5786-5798
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Biochemical characterization and crystal structure determination of human heart short chain L-3-hydroxyacyl-CoA dehydrogenase provide insights into catalytic mechanism.
|
|
J.J.Barycki,
L.K.O'Brien,
J.M.Bratt,
R.Zhang,
R.Sanishvili,
A.W.Strauss,
L.J.Banaszak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human heart short chain L-3-hydroxyacyl-CoA dehydrogenase (SCHAD) catalyzes the
oxidation of the hydroxyl group of L-3-hydroxyacyl-CoA to a keto group,
concomitant with the reduction of NAD+ to NADH, as part of the beta-oxidation
pathway. The homodimeric enzyme has been overexpressed in Escherichia coli,
purified to homogeneity, and studied using biochemical and crystallographic
techniques. The dissociation constants of NAD+ and NADH have been determined
over a broad pH range and indicate that SCHAD binds reduced cofactor
preferentially. Examination of apparent catalytic constants reveals that SCHAD
displays optimal enzymatic activity near neutral pH, with catalytic efficiency
diminishing rapidly toward pH extremes. The crystal structure of SCHAD complexed
with NAD+ has been solved using multiwavelength anomalous diffraction techniques
and a selenomethionine-substituted analogue of the enzyme. The subunit structure
is comprised of two domains. The first domain is similar to other alpha/beta
dinucleotide folds but includes an unusual helix-turn-helix motif which extends
from the central beta-sheet. The second, or C-terminal, domain is primarily
alpha-helical and mediates subunit dimerization and, presumably,
L-3-hydroxyacyl-CoA binding. Molecular modeling studies in which
L-3-hydroxybutyryl-CoA was docked into the enzyme-NAD+ complex suggest that His
158 serves as a general base, abstracting a proton from the 3-OH group of the
substrate. Furthermore, the ability of His 158 to perform such a function may be
enhanced by an electrostatic interaction with Glu 170, consistent with previous
biochemical observations. These studies provide further understanding of the
molecular basis of several inherited metabolic disease states correlated with
L-3-hydroxyacyl-CoA dehydrogenase deficiencies.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Parkot,
H.Gröger,
and
W.Hummel
(2010).
Purification, cloning, and overexpression of an alcohol dehydrogenase from Nocardia globerula reducing aliphatic ketones and bulky ketoesters.
|
| |
Appl Microbiol Biotechnol,
86,
1813-1820.
|
 |
|
|
|
|
 |
R.C.Taylor,
A.K.Brown,
A.Singh,
A.Bhatt,
and
G.S.Besra
(2010).
Characterization of a beta-hydroxybutyryl-CoA dehydrogenase from Mycobacterium tuberculosis.
|
| |
Microbiology,
156,
1975-1982.
|
 |
|
|
|
|
 |
N.Yokochi,
Y.Yoshikane,
S.Matsumoto,
M.Fujisawa,
K.Ohnishi,
and
T.Yagi
(2009).
Gene identification and characterization of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD+-dependent dismutase.
|
| |
J Biochem,
145,
493-503.
|
 |
|
|
|
|
 |
R.S.Rector,
R.M.Payne,
and
J.A.Ibdah
(2008).
Mitochondrial trifunctional protein defects: clinical implications and therapeutic approaches.
|
| |
Adv Drug Deliv Rev,
60,
1488-1496.
|
 |
|
|
|
|
 |
Y.Asada,
C.Kuroishi,
Y.Ukita,
R.Sumii,
S.Endo,
T.Matsunaga,
A.Hara,
and
N.Kunishima
(2008).
Crystallization and preliminary X-ray crystallographic analysis of rabbit L-gulonate 3-dehydrogenase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
228-230.
|
 |
|
|
|
|
 |
A.Ciulli,
D.Y.Chirgadze,
A.G.Smith,
T.L.Blundell,
and
C.Abell
(2007).
Crystal structure of Escherichia coli ketopantoate reductase in a ternary complex with NADP+ and pantoate bound: substrate recognition, conformational change, and cooperativity.
|
| |
J Biol Chem,
282,
8487-8497.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.S.Goetzman,
Y.Wang,
M.He,
A.W.Mohsen,
B.K.Ninness,
and
J.Vockley
(2007).
Expression and characterization of mutations in human very long-chain acyl-CoA dehydrogenase using a prokaryotic system.
|
| |
Mol Genet Metab,
91,
138-147.
|
 |
|
|
|
|
 |
W.Sun,
S.Singh,
R.Zhang,
J.L.Turnbull,
and
D.Christendat
(2006).
Crystal structure of prephenate dehydrogenase from Aquifex aeolicus. Insights into the catalytic mechanism.
|
| |
J Biol Chem,
281,
12919-12928.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Li,
T.Itoh,
T.Taguchi,
T.Xiang,
Y.Ebizuka,
and
K.Ichinose
(2005).
Functional studies on a ketoreductase gene from Streptomyces sp. AM-7161 to control the stereochemistry in medermycin biosynthesis.
|
| |
Bioorg Med Chem,
13,
6856-6863.
|
 |
|
|
|
|
 |
B.Nocek,
C.Chang,
H.Li,
L.Lezondra,
D.Holzle,
F.Collart,
and
A.Joachimiak
(2005).
Crystal structures of delta1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes.
|
| |
J Mol Biol,
354,
91.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Y.Yang,
X.Y.He,
and
H.Schulz
(2005).
3-Hydroxyacyl-CoA dehydrogenase and short chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease.
|
| |
FEBS J,
272,
4874-4883.
|
 |
|
|
|
|
 |
K.A.Lantz,
M.Z.Vatamaniuk,
J.E.Brestelli,
J.R.Friedman,
F.M.Matschinsky,
and
K.H.Kaestner
(2004).
Foxa2 regulates multiple pathways of insulin secretion.
|
| |
J Clin Invest,
114,
512-520.
|
 |
|
|
|
|
 |
M.Ishikawa,
D.Tsuchiya,
T.Oyama,
Y.Tsunaka,
and
K.Morikawa
(2004).
Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex.
|
| |
EMBO J,
23,
2745-2754.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.W.Aufhammer,
E.Warkentin,
H.Berk,
S.Shima,
R.K.Thauer,
and
U.Ermler
(2004).
Coenzyme binding in F420-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family.
|
| |
Structure,
12,
361-370.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
U.Spiekerkoetter,
Z.Khuchua,
Z.Yue,
M.J.Bennett,
and
A.W.Strauss
(2004).
General mitochondrial trifunctional protein (TFP) deficiency as a result of either alpha- or beta-subunit mutations exhibits similar phenotypes because mutations in either subunit alter TFP complex expression and subunit turnover.
|
| |
Pediatr Res,
55,
190-196.
|
 |
|
|
|
|
 |
J.P.Taskinen,
T.R.Kiema,
K.T.Koivuranta,
R.K.Wierenga,
and
J.K.Hiltunen
(2002).
Crystallization and characterization of the dehydrogenase domain from rat peroxisomal multifunctional enzyme type 1.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
690-693.
|
 |
|
|
|
|
 |
J.Vockley,
R.H.Singh,
and
D.A.Whiteman
(2002).
Diagnosis and management of defects of mitochondrial beta-oxidation.
|
| |
Curr Opin Clin Nutr Metab Care,
5,
601-609.
|
 |
|
|
|
|
 |
D.G.Levitt
(2001).
A new software routine that automates the fitting of protein X-ray crystallographic electron-density maps.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1013-1019.
|
 |
|
|
|
|
 |
E.Warkentin,
B.Mamat,
M.Sordel-Klippert,
M.Wicke,
R.K.Thauer,
M.Iwata,
S.Iwata,
U.Ermler,
and
S.Shima
(2001).
Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
|
| |
EMBO J,
20,
6561-6569.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.T.Clayton,
S.Eaton,
A.Aynsley-Green,
M.Edginton,
K.Hussain,
S.Krywawych,
V.Datta,
H.E.Malingre,
R.Berger,
and
I.E.van den Berg
(2001).
Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion.
|
| |
J Clin Invest,
108,
457-465.
|
 |
|
|
|
|
 |
R.E.Campbell,
S.C.Mosimann,
I.van De Rijn,
M.E.Tanner,
and
N.C.Strynadka
(2000).
The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
|
| |
Biochemistry,
39,
7012-7023.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.J.Barycki,
L.K.O'Brien,
J.J.Birktoft,
A.W.Strauss,
and
L.J.Banaszak
(1999).
Pig heart short chain L-3-hydroxyacyl-CoA dehydrogenase revisited: sequence analysis and crystal structure determination.
|
| |
Protein Sci,
8,
2010-2018.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.A.Walsh,
G.Evans,
R.Sanishvili,
I.Dementieva,
and
A.Joachimiak
(1999).
MAD data collection - current trends.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1726-1732.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

