 |
PDBsum entry 2h9c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.99.21
- isochorismate lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isochorismate = salicylate + pyruvate
|
 |
 |
 |
 |
 |
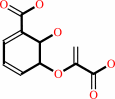 isochorismate
isochorismate
|
=
|
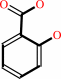 salicylate
salicylate
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.99.5
- chorismate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = prephenate
|
 |
 |
 |
 |
 |
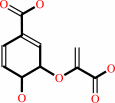 chorismate
chorismate
|
=
|
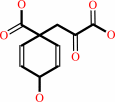 prephenate
prephenate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:33441-33449
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Two crystal structures of the isochorismate pyruvate lyase from Pseudomonas aeruginosa.
|
|
J.Zaitseva,
J.Lu,
K.L.Olechoski,
A.L.Lamb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Enzymatic systems that exploit pericyclic reaction mechanisms are rare. A recent
addition to this class is the enzyme PchB, an 11.4-kDa isochorismate pyruvate
lyase from Pseudomonas aeruginosa. The apo and pyruvate-bound structures of PchB
reveal that the enzyme is a structural homologue of chorismate mutases in the
AroQalpha class despite low sequence identity (20%). The enzyme is an
intertwined dimer of three helices with connecting loops, and amino acids from
each monomer participate in each of two active sites. The apo structure (2.35 A
resolution) has one dimer per asymmetric unit with nitrate bound in an open
active site. The loop between the first and second helices is disordered,
providing a gateway for substrate entry and product exit. The pyruvate-bound
structure (1.95 A resolution) has two dimers per asymmetric unit. One has two
open active sites like the apo structure, and the other has two closed active
sites with the loop between the first and second helices ordered for catalysis.
Determining the structure of PchB is part of a larger effort to elucidate
protein structures involved in siderophore biosynthesis, as these enzymes are
crucial for bacterial iron uptake and virulence and have been identified as
antimicrobial drug targets.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
FIGURE 5. Stereoviews of the active sites of apo PchB (a)
with the closed pyruvate-bound PchB (b) and EcCM (c). The colors
used are as described in Fig. 2. The amino acid legends in panel
b indicate monomer name, residue number, and one-letter amino
acid code and are in capital letters if they are conserved
between PchB and EcCM and lowercase if they are not.
|
 |
Figure 6.
FIGURE 6. Two-dimensional schematic representation of the
protein-ligand contacts in the closed, pyruvate-bound structure
(a) and the wrong-ligand crystallographic refinement model
containing salicylate and pyruvate (b). The amino acid side
chains are shown as sticks and labeled with the three-letter
amino acid code, amino acid number, and monomer name. The
pyruvate and salicylate molecules are depicted as ball-and-stick
and are labeled "Pyr 1," "Pyr 2," and "SalI" with a monomer name
of (E). The carbon atoms are black, and the oxygen atoms are
white. This figure was generated with LIGPLOT (31).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
33441-33449)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.A.Youard,
N.Wenner,
and
C.Reimmann
(2011).
Iron acquisition with the natural siderophore enantiomers pyochelin and enantio-pyochelin in Pseudomonas species.
|
| |
Biometals,
24,
513-522.
|
 |
|
|
|
|
 |
C.Jäckel,
P.Kast,
and
D.Hilvert
(2008).
Protein design by directed evolution.
|
| |
Annu Rev Biophys,
37,
153-173.
|
 |
|
|
|
|
 |
O.Kerbarh,
A.Ciulli,
D.Y.Chirgadze,
T.L.Blundell,
and
C.Abell
(2007).
Nucleophile selectivity of chorismate-utilizing enzymes.
|
| |
Chembiochem,
8,
622-624.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

