 |
PDBsum entry 2h7c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of human carboxylesterase in complex with coenzyme a
|
|
Structure:
|
 |
Liver carboxylesterase 1. Chain: a, b, c, d, e, f. Fragment: residues 19-561. Synonym: acyl coenzyme a:cholesterol acyltransferase, acat, monocyte/macrophage serine esterase, hmse, serine esterase 1, brain carboxylesterase hbr1, triacylglycerol hydrolase, tgh, egasyn. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Expression_system_cell_line: sf9.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.183
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
S.Bencharit,C.C.Edwards,C.L.Morton,E.L.Howard-Williams,P.M.Potter, M.R.Redinbo
|
Key ref:

|
 |
S.Bencharit
et al.
(2006).
Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1.
J Mol Biol,
363,
201-214.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Jun-06
|
Release date:
|
29-Aug-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P23141
(EST1_HUMAN) -
Liver carboxylesterase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
567 a.a.
532 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.1.1
- carboxylesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a carboxylic ester + H2O = an alcohol + a carboxylate + H+
|
 |
 |
 |
 |
 |
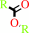 carboxylic ester
carboxylic ester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
 carboxylate
carboxylate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.1.13
- sterol esterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a sterol ester + H2O = a sterol + a fatty acid + H+
|
 |
 |
 |
 |
 |
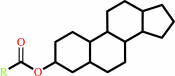 sterol ester
sterol ester
|
+
|
H2O
|
=
|
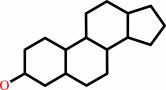 sterol
sterol
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.1.56
- methylumbelliferyl-acetate deacetylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-methylumbelliferyl acetate + H2O = 4-methylumbelliferone + acetate + H+
|
 |
 |
 |
 |
 |
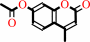 4-methylumbelliferyl acetate
4-methylumbelliferyl acetate
|
+
|
H2O
|
=
|
4-methylumbelliferone
Bound ligand (Het Group name = )
matches with 54.55% similarity
|
+
|
 acetate
acetate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
363:201-214
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1.
|
|
S.Bencharit,
C.C.Edwards,
C.L.Morton,
E.L.Howard-Williams,
P.Kuhn,
P.M.Potter,
M.R.Redinbo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human carboxylesterase 1 (hCE1) is a drug and endobiotic-processing serine
hydrolase that exhibits relatively broad substrate specificity. It has been
implicated in a variety of endogenous cholesterol metabolism pathways including
the following apparently disparate reactions: cholesterol ester hydrolysis
(CEH), fatty acyl Coenzyme A hydrolysis (FACoAH), acyl-Coenzyme A:cholesterol
acyltransfer (ACAT), and fatty acyl ethyl ester synthesis (FAEES). The
structural basis for the ability of hCE1 to perform these catalytic actions
involving large substrates and products has remained unclear. Here we present
four crystal structures of the hCE1 glycoprotein in complexes with the following
endogenous substrates or substrate analogues: Coenzyme A, the fatty acid
palmitate, and the bile acids cholate and taurocholate. While the active site of
hCE1 was known to be promiscuous and capable of interacting with a variety of
chemically distinct ligands, these structures reveal that the enzyme contains
two additional ligand-binding sites and that each site also exhibits relatively
non-specific ligand-binding properties. Using this multisite promiscuity, hCE1
appears structurally capable of assembling several catalytic events depending,
apparently, on the physiological state of the cellular environment. These
results expand our understanding of enzyme promiscuity and indicate that, in the
case of hCE1, multiple non-specific sites are employed to perform distinct
catalytic actions.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Elsevier:
J Mol Biol
(2006,
363,
201-214)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Li,
J.E.Janecka,
and
W.J.Murphy
(2011).
Accelerated evolution of CES7, a gene encoding a novel major urinary protein in the cat family.
|
| |
Mol Biol Evol,
28,
911-920.
|
 |
|
|
|
|
 |
G.Vistoli,
A.Pedretti,
A.Mazzolari,
and
B.Testa
(2010).
Homology modeling and metabolism prediction of human carboxylesterase-2 using docking analyses by GriDock: a parallelized tool based on AutoDock 4.0.
|
| |
J Comput Aided Mol Des,
24,
771-787.
|
 |
|
|
|
|
 |
J.A.Crow,
K.L.Herring,
S.Xie,
A.Borazjani,
P.M.Potter,
and
M.K.Ross
(2010).
Inhibition of carboxylesterase activity of THP1 monocytes/macrophages and recombinant human carboxylesterase 1 by oxysterols and fatty acids.
|
| |
Biochim Biophys Acta,
1801,
31-41.
|
 |
|
|
|
|
 |
J.Rayo,
L.Muñoz,
G.Rosell,
B.D.Hammock,
A.Guerrero,
F.J.Luque,
and
R.Pouplana
(2010).
Reactivity versus steric effects in fluorinated ketones as esterase inhibitors: a quantum mechanical and molecular dynamics study.
|
| |
J Mol Model,
16,
1753-1764.
|
 |
|
|
|
|
 |
R.S.Holmes,
L.A.Cox,
and
J.L.VandeBerg
(2010).
Mammalian carboxylesterase 3: comparative genomics and proteomics.
|
| |
Genetica,
138,
695-708.
|
 |
|
|
|
|
 |
R.S.Holmes,
M.W.Wright,
S.J.Laulederkind,
L.A.Cox,
M.Hosokawa,
T.Imai,
S.Ishibashi,
R.Lehner,
M.Miyazaki,
E.J.Perkins,
P.M.Potter,
M.R.Redinbo,
J.Robert,
T.Satoh,
T.Yamashita,
B.Yan,
T.Yokoi,
R.Zechner,
and
L.J.Maltais
(2010).
Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins.
|
| |
Mamm Genome,
21,
427-441.
|
 |
|
|
|
|
 |
G.A.Mitchell
(2009).
Genetics, physiology and perinatal influences in childhood obesity: view from the Chair.
|
| |
Int J Obes (Lond),
33,
S41-S47.
|
 |
|
|
|
|
 |
M.Jernås,
B.Olsson,
P.Arner,
P.Jacobson,
L.Sjöström,
A.Walley,
P.Froguel,
P.G.McTernan,
J.Hoffstedt,
and
L.M.Carlsson
(2009).
Regulation of carboxylesterase 1 (CES1) in human adipose tissue.
|
| |
Biochem Biophys Res Commun,
383,
63-67.
|
 |
|
|
|
|
 |
N.S.Lamango,
R.Duverna,
W.Zhang,
and
S.Y.Ablordeppey
(2009).
Porcine Liver Carboxylesterase Requires Polyisoprenylation for High Affinity Binding to Cysteinyl Substrates.
|
| |
Open Enzym Inhib J,
2,
12-27.
|
 |
|
|
|
|
 |
R.S.Holmes,
J.P.Glenn,
J.L.VandeBerg,
and
L.A.Cox
(2009).
Baboon carboxylesterases 1 and 2: sequences, structures and phylogenetic relationships with human and other primate carboxylesterases.
|
| |
J Med Primatol,
38,
27-38.
|
 |
|
|
|
|
 |
R.S.Holmes,
L.A.Cox,
and
J.L.Vandeberg
(2009).
A new class of mammalian carboxylesterase CES6.
|
| |
Comp Biochem Physiol Part D Genomics Proteomics,
4,
209-217.
|
 |
|
|
|
|
 |
R.S.Holmes,
L.A.Cox,
and
J.L.Vandeberg
(2009).
Bovine Carboxylesterases: Evidence for Two CES1 and Five Families of CES Genes on Chromosome 18.
|
| |
Comp Biochem Physiol Part D Genomics Proteomics,
4,
11-20.
|
 |
|
|
|
|
 |
R.S.Holmes,
L.A.Cox,
and
J.L.Vandeberg
(2009).
Horse carboxylesterases: evidence for six CES1 and four families of CES genes on chromosome 3.
|
| |
Comp Biochem Physiol Part D Genomics Proteomics,
4,
54-65.
|
 |
|
|
|
|
 |
T.Harada,
Y.Nakagawa,
R.M.Wadkins,
P.M.Potter,
and
C.E.Wheelock
(2009).
Comparison of benzil and trifluoromethyl ketone (TFK)-mediated carboxylesterase inhibition using classical and 3D-quantitative structure-activity relationship analysis.
|
| |
Bioorg Med Chem,
17,
149-164.
|
 |
|
|
|
|
 |
J.A.Crow,
B.L.Middleton,
A.Borazjani,
M.J.Hatfield,
P.M.Potter,
and
M.K.Ross
(2008).
Inhibition of carboxylesterase 1 is associated with cholesteryl ester retention in human THP-1 monocyte/macrophages.
|
| |
Biochim Biophys Acta,
1781,
643-654.
|
 |
|
|
|
|
 |
R.S.Holmes,
J.Chan,
L.A.Cox,
W.J.Murphy,
and
J.L.VandeBerg
(2008).
Opossum carboxylesterases: sequences, phylogeny and evidence for CES gene duplication events predating the marsupial-eutherian common ancestor.
|
| |
BMC Evol Biol,
8,
54.
|
 |
|
|
|
|
 |
R.S.Holmes,
L.A.Cox,
and
J.L.Vandeberg
(2008).
Mammalian carboxylesterase 5: comparative biochemistry and genomics.
|
| |
Comp Biochem Physiol Part D Genomics Proteomics,
3,
195-204.
|
 |
|
|
|
|
 |
S.Takahashi,
M.Katoh,
T.Saitoh,
M.Nakajima,
and
T.Yokoi
(2008).
Allosteric kinetics of human carboxylesterase 1: species differences and interindividual variability.
|
| |
J Pharm Sci,
97,
5434-5445.
|
 |
|
|
|
|
 |
T.M.Streit,
A.Borazjani,
S.E.Lentz,
M.Wierdl,
P.M.Potter,
S.R.Gwaltney,
and
M.K.Ross
(2008).
Evaluation of the 'side door' in carboxylesterase-mediated catalysis and inhibition.
|
| |
Biol Chem,
389,
149-162.
|
 |
|
|
|
|
 |
C.D.Fleming,
C.C.Edwards,
S.D.Kirby,
D.M.Maxwell,
P.M.Potter,
D.M.Cerasoli,
and
M.R.Redinbo
(2007).
Crystal structures of human carboxylesterase 1 in covalent complexes with the chemical warfare agents soman and tabun.
|
| |
Biochemistry,
46,
5063-5071.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Liu,
H.E.Ewis,
P.C.Tai,
C.D.Lu,
and
I.T.Weber
(2007).
Crystal structure of the Geobacillus stearothermophilus carboxylesterase Est55 and its activation of prodrug CPT-11.
|
| |
J Mol Biol,
367,
212-223.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

