 |
PDBsum entry 2h4x

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Human bisphosphoglycerate mutase complex with 3-phosphoglycerate with crystal growth 90 days
|
|
Structure:
|
 |
Bisphosphoglycerate mutase. Chain: a, b. Synonym: 2,3-bisphosphoglycerate mutase, erythrocyte, 2,3- bisphosphoglycerate synthase, bpgm, bpg-dependent pgam. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
1.85Å
|
R-factor:
|
0.180
|
R-free:
|
0.215
|
|
|
Authors:
|
 |
Y.Wang,W.Gong
|
Key ref:

|
 |
Y.Wang
et al.
(2006).
Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
J Biol Chem,
281,
39642-39648.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
25-May-06
|
Release date:
|
24-Oct-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07738
(PMGE_HUMAN) -
Bisphosphoglycerate mutase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
259 a.a.
255 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.4.2.11
- phosphoglycerate mutase (2,3-diphosphoglycerate-dependent).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R)-2-phosphoglycerate = (2R)-3-phosphoglycerate
|
 |
 |
 |
 |
 |
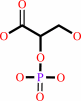 2-phospho-D-glycerate
2-phospho-D-glycerate
|
=
|
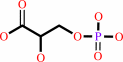 3-phospho-D-glycerate
3-phospho-D-glycerate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.2.4
- bisphosphoglycerate mutase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R)-3-phospho-glyceroyl phosphate = (2R)-2,3-bisphosphoglycerate + H+
|
 |
 |
 |
 |
 |
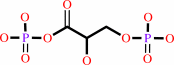 3-phospho-D-glyceroyl phosphate
3-phospho-D-glyceroyl phosphate
|
=
|
 2,3-bisphospho-D-glycerate
2,3-bisphospho-D-glycerate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:39642-39648
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
|
|
Y.Wang,
L.Liu,
Z.Wei,
Z.Cheng,
Y.Lin,
W.Gong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Bisphosphoglycerate mutase is an erythrocyte-specific enzyme catalyzing a series
of intermolecular phosphoryl group transfer reactions. Its main function is to
synthesize 2,3-bisphosphoglycerate, the allosteric effector of hemoglobin. In
this paper, we directly observed real-time motion of the enzyme active site and
the substrate during phosphoryl transfer. A series of high resolution crystal
structures of human bisphosphoglycerate mutase co-crystallized with
2,3-bisphosphoglycerate, representing different time points in the phosphoryl
transfer reaction, were solved. These structures not only clarify the argument
concerning the substrate binding mode for this enzyme family but also depict the
entire process of the key histidine phosphorylation as a "slow movie".
It was observed that the enzyme conformation continuously changed during the
different states of the reaction. These results provide direct evidence for an
"in line" phosphoryl transfer mechanism, and the roles of some key
residues in the phosphoryl transfer process are identified.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. A F[o] - F[c] electron density map (contoured at
3.5  , purple cage) at the
active site when the side chain of His-11 and the ligand are
omitted. A (structure 1), co-crystallized with 2,3-BPG at 4
°C. B (structure 2), co-crystallized with 2,3-BPG at 16
°C for less than 16 days. C (structure 3), only 3-PGA
binding. D (structure 4), co-crystallized with 2,3-BPG at 16
°C for 17 days. Extra F[o] - F[c] electron density
(contoured at 3 , purple cage) at the
active site when the side chain of His-11 and the ligand are
omitted. A (structure 1), co-crystallized with 2,3-BPG at 4
°C. B (structure 2), co-crystallized with 2,3-BPG at 16
°C for less than 16 days. C (structure 3), only 3-PGA
binding. D (structure 4), co-crystallized with 2,3-BPG at 16
°C for 17 days. Extra F[o] - F[c] electron density
(contoured at 3  , blue cage) exists
when two water molecules and 3-PGA are modeled in. E (structure
5), co-crystallized with , blue cage) exists
when two water molecules and 3-PGA are modeled in. E (structure
5), co-crystallized with  and
3-PGA. and
3-PGA.
|
 |
Figure 4.
FIGURE 4. Schematic of the interactions between the ligand
and human BPGM with all interactions between the ligand and
residues depicted as dashed lines and labeled with the bond
lengths (Å). A, interactions between 2,3-BPG and BPGM in
the reacting state. B, interaction between 3-PGA and the
phosphorylated enzyme.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
39642-39648)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Chen,
J.Jakoncic,
K.A.Parker,
N.Carpino,
and
N.Nassar
(2009).
Structures of the phosphorylated and VO(3)-bound 2H-phosphatase domain of Sts-2.
|
| |
Biochemistry,
48,
8129-8135.
|
 |
|
|
|
|
 |
Z.Lu,
D.Dunaway-Mariano,
and
K.N.Allen
(2008).
The catalytic scaffold of the haloalkanoic acid dehalogenase enzyme superfamily acts as a mold for the trigonal bipyramidal transition state.
|
| |
Proc Natl Acad Sci U S A,
105,
5687-5692.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

