 |
PDBsum entry 2ger

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2ger
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure and oxidative mechanism of human pyrroline-5- carboxylate reductase
|
|
Structure:
|
 |
Pyrroline-5-carboxylate reductase 1. Chain: a, b, c, d, e. Synonym: p5cr 1, p5c reductase 1. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
3.10Å
|
R-factor:
|
0.233
|
R-free:
|
0.261
|
|
|
Authors:
|
 |
Z.Meng,Z.Lou,Z.Liu,Z.Rao
|
Key ref:

|
 |
Z.Meng
et al.
(2006).
Crystal structure of human pyrroline-5-carboxylate reductase.
J Mol Biol,
359,
1364-1377.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
20-Mar-06
|
Release date:
|
19-Sep-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P32322
(P5CR1_HUMAN) -
Pyrroline-5-carboxylate reductase 1, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
319 a.a.
277 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.5.1.2
- pyrroline-5-carboxylate reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Proline Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-proline + NAD+ = (S)-1-pyrroline-5-carboxylate + NADH + 2 H+
|
|
2.
|
L-proline + NADP+ = (S)-1-pyrroline-5-carboxylate + NADPH + 2 H+
|
|
 |
 |
 |
 |
 |
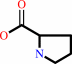 L-proline
L-proline
|
+
|
 NAD(+)
NAD(+)
|
=
|
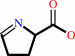 (S)-1-pyrroline-5-carboxylate
(S)-1-pyrroline-5-carboxylate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 L-proline
L-proline
|
+
|
 NADP(+)
NADP(+)
|
=
|
 (S)-1-pyrroline-5-carboxylate
(S)-1-pyrroline-5-carboxylate
|
+
|
 NADPH
NADPH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
359:1364-1377
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human pyrroline-5-carboxylate reductase.
|
|
Z.Meng,
Z.Lou,
Z.Liu,
M.Li,
X.Zhao,
M.Bartlam,
Z.Rao.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Pyrroline-5-carboxylate reductase (P5CR) is a universal housekeeping enzyme that
catalyzes the reduction of Delta(1)-pyrroline-5-carboxylate (P5C) to proline
using NAD(P)H as the cofactor. The enzymatic cycle between P5C and proline is
very important for the regulation of amino acid metabolism, intracellular redox
potential, and apoptosis. Here, we present the 2.8 Angstroms resolution
structure of the P5CR apo enzyme, its 3.1 Angstroms resolution ternary complex
with NAD(P)H and substrate-analog. The refined structures demonstrate a
decameric architecture with five homodimer subunits and ten catalytic sites
arranged around a peripheral circular groove. Mutagenesis and kinetic studies
reveal the pivotal roles of the dinucleotide-binding Rossmann motif and residue
Glu221 in the human enzyme. Human P5CR is thermostable and the crystals were
grown at 37 degrees C. The enzyme is implicated in oxidation of the anti-tumor
drug thioproline.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The P5CR structure. (a) Overall view of the P5CR
crystal structure. A ribbon representation of only one homodimer
is shown, with each monomer presented in magenta and gold,
respectively. The other four homodimers, which form the decamer,
are shown in molecular surface representation. (b) The potential
surface of the decamer ternary complex. The binding cofactor and
substrate analog are shown as magenta spheres around the
circular groove. The three dimensions are labeled and the
negatively charged central channel with 25 Å diameter is
shown. (c) Two views related by 90° of the monomer structure
of P5CR, showing domains A and B with their secondary structure
units in detail. (d) Two views related by 90° of the dimer
structure of P5CR, showing how two monomers coil around each
other to form one dimer. Monomers are shown in cartoon
representation in magenta and gold, respectively. Secondary
structure units of molecule 1 are named as αx-1; and in
molecule 2 are named as αx-2. (e) Stereo view of the
inter-homodimer interactions. Only two homodimers are shown at
the interface. Molecules A and H, which belong to one dimer, are
shown in magenta and yellow, respectively; molecules E and I,
which belong to the other dimer, are drawn in green and red,
respectively. There are 11 inter-homodimer, five water-mediated
inter-molecular and four inntra-molecular hydrogen bonds, which
form an interaction web between two homodimers. Hydrogen bonds
are shown as broken cyan lines. (f) Investigation of the decamer
stability with different concentrations of urea. In the
Superdex200 profiles, P5CR eluted at the decamer peak when
treated with no urea (black) or with 0.5 M urea (magenta). The
decamer began to dissociate into dimers from 1 M urea (yellow)
and 2 M urea (blue) and dissociated completely into dimers at 4
M urea (red). P5CR was refolded into a decamer from 6 M urea in
Superdex200 (shown as a green curve). No monomer was observed
for any concentration of urea. (g) Thermal inactivation of P5CR:
(1) after incubation at various temperatures (20–75 °C)
for 10 min, the relative activities of P5CR were measured at 25
°C. (2) The time-dependent thermal inactivation of P5CR was
investigated at 40 °C and at 68 °C. The relative
activities were measured at 25 °C and plotted against time.
|
 |
Figure 3.
Figure 3. (a) Structure-based sequence alignment between
P5CR in human and in different organisms as indicated. Arrows
indicate β-strands; cylinders denote α-helices. Background-red
residues indicate those that are conserved; background-yellow
denotes residues identified to be more than 80% conserved.
Residues that are important for cofactor and substrate analog
binding are framed in black. (b) Superposition of P5CR from
human (yellow), Neisseria meningitides Mc58 (green) and
Streptococcus pyogenes (red). The α2 helix of all three
structures is shown in ribbon representation and the other parts
are shown as smooth lines. The NADP cofactor bound in the
structure of P5CR from Streptococcus pyogenes is shown in
magenta stick representation.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
359,
1364-1377)
copyright 2006.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Phang,
W.Liu,
and
O.Zabirnyk
(2010).
Proline metabolism and microenvironmental stress.
|
| |
Annu Rev Nutr,
30,
441-463.
|
 |
|
|
|
|
 |
B.Reversade,
N.Escande-Beillard,
A.Dimopoulou,
B.Fischer,
S.C.Chng,
Y.Li,
M.Shboul,
P.Y.Tham,
H.Kayserili,
L.Al-Gazali,
M.Shahwan,
F.Brancati,
H.Lee,
B.D.O'Connor,
M.Schmidt-von Kegler,
B.Merriman,
S.F.Nelson,
A.Masri,
F.Alkazaleh,
D.Guerra,
P.Ferrari,
A.Nanda,
A.Rajab,
D.Markie,
M.Gray,
J.Nelson,
A.Grix,
A.Sommer,
R.Savarirayan,
A.R.Janecke,
E.Steichen,
D.Sillence,
I.Hausser,
B.Budde,
G.Nürnberg,
P.Nürnberg,
P.Seemann,
D.Kunkel,
G.Zambruno,
B.Dallapiccola,
M.Schuelke,
S.Robertson,
H.Hamamy,
B.Wollnik,
L.Van Maldergem,
S.Mundlos,
and
U.Kornak
(2009).
Mutations in PYCR1 cause cutis laxa with progeroid features.
|
| |
Nat Genet,
41,
1016-1021.
|
 |
|
|
|
|
 |
Y.Ma,
Z.Ding,
Y.Qian,
Y.W.Wan,
K.Tosun,
X.Shi,
V.Castranova,
E.J.Harner,
and
N.L.Guo
(2009).
An integrative genomic and proteomic approach to chemosensitivity prediction.
|
| |
Int J Oncol,
34,
107-115.
|
 |
|
|
|
|
 |
C.A.Hu,
D.Bart Williams,
S.Zhaorigetu,
S.Khalil,
G.Wan,
and
D.Valle
(2008).
Functional genomics and SNP analysis of human genes encoding proline metabolic enzymes.
|
| |
Amino Acids,
35,
655-664.
|
 |
|
|
|
|
 |
J.J.Tanner
(2008).
Structural biology of proline catabolism.
|
| |
Amino Acids,
35,
719-730.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

