 |
PDBsum entry 2g7f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:1387-1391
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion.
|
|
B.Altermark,
A.O.Smalås,
N.P.Willassen,
R.Helland.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of a periplasmic/extracellular endonuclease from Vibrio
cholerae has been solved at low and at neutral pH. Crystals grown at pH 4.6 and
6.9 diffracted to 1.6 A (on BM01A at the ESRF) and 1.95 A (on a rotating-anode
generator), respectively. The structures of the endonuclease were compared with
the structure of a homologous enzyme in V. vulnificus. The structures of the V.
cholerae enzyme at different pH values are essentially identical to each other
and to the V. vulnificus enzyme. However, interesting features were observed in
the solvent structures. Both V. cholerae structures reveal the presence of a
chloride ion completely buried within the core of the protein, with the nearest
solvent molecule approximately 7 A away. Magnesium, which is essential for
catalysis, is present in the structure at neutral pH, but is absent at low pH,
and may partly explain the inactivity of the enzyme at lower pH.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 2.
Figure 2 (a) Stereo plot illustrating the anomalous difference
map (red) contoured at 4.5  surrounding
the buried chloride (green) in the interior of the protein and
the S atoms (yellow). Magnesium is displayed as an orange
sphere. (b) Stereo plot of the interactions between the chloride
ion (green sphere) and the protein main-chain N atoms of Tyr43,
Cys44, Ile123 and the O^ surrounding
the buried chloride (green) in the interior of the protein and
the S atoms (yellow). Magnesium is displayed as an orange
sphere. (b) Stereo plot of the interactions between the chloride
ion (green sphere) and the protein main-chain N atoms of Tyr43,
Cys44, Ile123 and the O^ 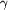 atom
of Ser41. Two disulfide bridges in the vicinity of the
chloride-binding site are also displayed. This figure was
prepared with PyMOL (DeLano, 2002[DeLano, W. L. (2002). The
PyMOL Molecular Visualization System. http://www.pymol.org .])
and MOLSCRIPT (Kraulis, 1991[Kraulis, P. J. (1991). J. Appl.
Cryst. 24, 946-950.]). atom
of Ser41. Two disulfide bridges in the vicinity of the
chloride-binding site are also displayed. This figure was
prepared with PyMOL (DeLano, 2002[DeLano, W. L. (2002). The
PyMOL Molecular Visualization System. http://www.pymol.org .])
and MOLSCRIPT (Kraulis, 1991[Kraulis, P. J. (1991). J. Appl.
Cryst. 24, 946-950.]).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
1387-1391)
copyright 2006.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.Yang
(2011).
Nucleases: diversity of structure, function and mechanism.
|
| |
Q Rev Biophys,
44,
1.
|
 |
|
|
|
|
 |
D.Vertommen,
M.Depuydt,
J.Pan,
P.Leverrier,
L.Knoops,
J.P.Szikora,
J.Messens,
J.C.Bardwell,
and
J.F.Collet
(2008).
The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner.
|
| |
Mol Microbiol,
67,
336-349.
|
 |
|
|
|
|
 |
L.Niiranen,
B.Altermark,
B.O.Brandsdal,
H.K.Leiros,
R.Helland,
A.O.Smalås,
and
N.P.Willassen
(2008).
Effects of salt on the kinetics and thermodynamic stability of endonuclease I from Vibrio salmonicida and Vibrio cholerae.
|
| |
FEBS J,
275,
1593-1605.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Blokesch,
and
G.K.Schoolnik
(2008).
The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae.
|
| |
J Bacteriol,
190,
7232-7240.
|
 |
|
|
|
|
 |
B.Altermark,
L.Niiranen,
N.P.Willassen,
A.O.Smalås,
and
E.Moe
(2007).
Comparative studies of endonuclease I from cold-adapted Vibrio salmonicida and mesophilic Vibrio cholerae.
|
| |
FEBS J,
274,
252-263.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

