 |
PDBsum entry 2g6c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.9
- N-acetylornithine carbamoyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N2-acetyl-L-ornithine + carbamoyl phosphate = N2-acetyl-L-citrulline + phosphate + H+
|
 |
 |
 |
 |
 |
N(2)-acetyl-L-ornithine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
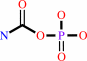 carbamoyl phosphate
carbamoyl phosphate
|
=
|
N(2)-acetyl-L-citrulline
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
16:1689-1699
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
|
|
D.Shi,
X.Yu,
J.Cabrera-Luque,
T.Y.Chen,
L.Roth,
H.Morizono,
N.M.Allewell,
M.Tuchman.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Transcarbamylases catalyze the transfer of the carbamyl group from carbamyl
phosphate (CP) to an amino group of a second substrate such as aspartate,
ornithine, or putrescine. Previously, structural determination of a
transcarbamylase from Xanthomonas campestris led to the discovery of a novel
N-acetylornithine transcarbamylase (AOTCase) that catalyzes the carbamylation of
N-acetylornithine. Recently, a novel N-succinylornithine transcarbamylase
(SOTCase) from Bacteroides fragilis was identified. Structural comparisons of
AOTCase from X. campestris and SOTCase from B. fragilis revealed that residue
Glu92 (X. campestris numbering) plays a critical role in distinguishing AOTCase
from SOTCase. Enzymatic assays of E92P, E92S, E92V, and E92A mutants of AOTCase
demonstrate that each of these mutations converts the AOTCase to an SOTCase.
Similarly, the P90E mutation in B. fragilis SOTCase (equivalent to E92 in X.
campestris AOTCase) converts the SOTCase to AOTCase. Hence, a single amino acid
substitution is sufficient to swap the substrate specificities of AOTCase and
SOTCase. X-ray crystal structures of these mutants in complexes with CP and
N-acetyl-L-norvaline (an analog of N-acetyl-L-ornithine) or
N-succinyl-L-norvaline (an analog of N-succinyl-L-ornithine) substantiate this
conversion. In addition to Glu92 (X. campestris numbering), other residues such
as Asn185 and Lys30 in AOTCase, which are involved in binding substrates through
bridging water molecules, help to define the substrate specificity of AOTCase.
These results provide the correct annotation (AOTCase or SOTCase) for a set of
the transcarbamylase-like proteins that have been erroneously annotated as
ornithine transcarbamylase (OTCase, EC 2.1.3.3).

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. N-acetylnorvaline or N-succinylnorvaline binding of SOTCase or AOTCase mutants. (A--D) Contours of the electron density
|
 |
Figure 5.
Figure 5. A schematic diagram representing the classification of four
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2007,
16,
1689-1699)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

